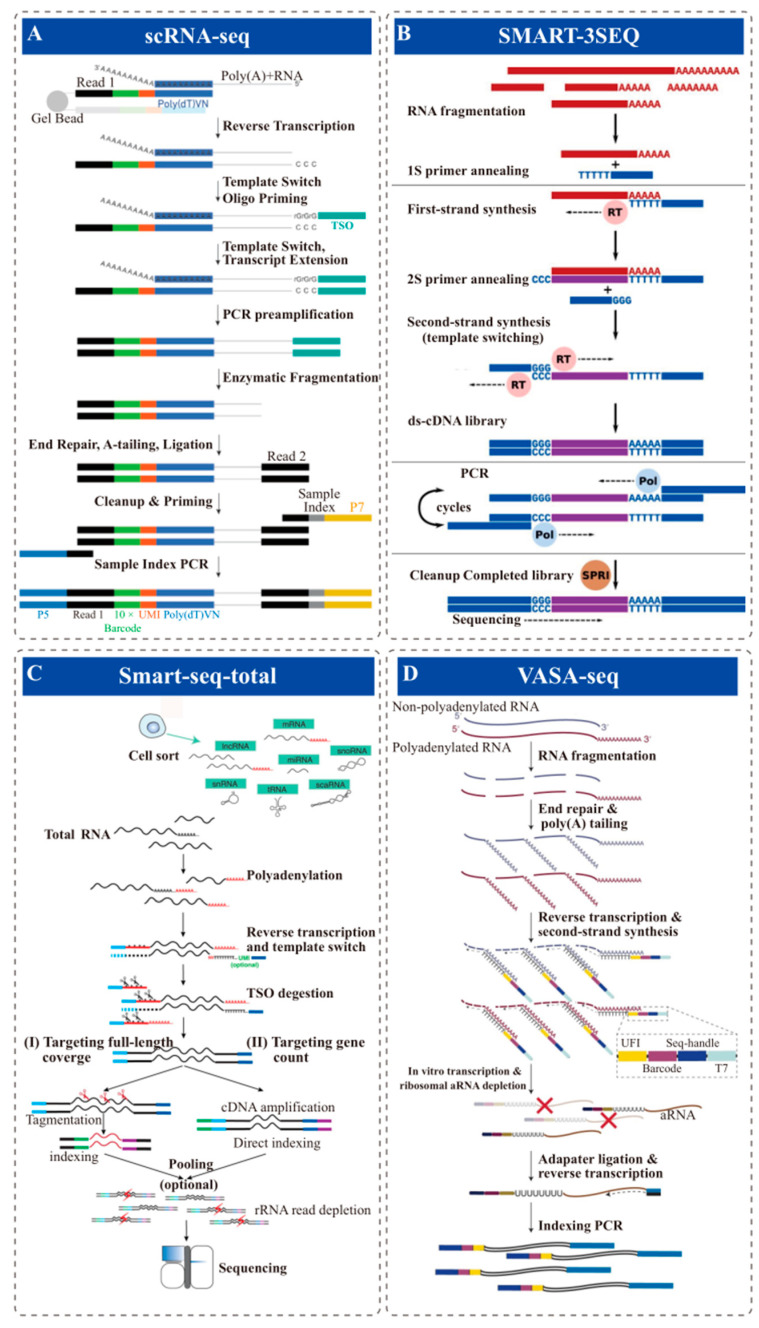
Figure 1.
sc/snRNA-seq of FFPE samples based on Oligo-dT probe capture techniques. (A) Traditional 3′ sequencing library construction principle (ex., 10×, drop-seq, AccuraCell™). (B) Conceptual diagram of the Smart-3SEQ library preparation method. Reprinted with permission from ref. [31]. Copyright 2019 Genome Research. RNA is denatured and fragmented by hydrolysis; The oligo(dT) primer captures poly-dA; Synthesizes first-strand cDNA and the second cDNA strand; PCR with long primers amplifies and cleanup step uses SPRI beads; The final library contains the unknown cDNA sequence between the two sequencing adapters. (C) Schematic of Smart-seq-total pipelines. Reprinted with permission from ref. [32]. Copyright 2021 PNAS. Cell lysis, total RNA is polyadenylated, primed with anchored oligo-dT; After reverse transcription, TSO is enzymatically cleaved, single-stranded cDNA is amplified and cleaned up; Amplified cDNA is then either tagmented or directly indexed, pooled, and depleted from ribosomal sequences. (D) Overview of the VASA-seq single-cell molecular workflow. Reprinted with permission from ref. [36]. Copyright 2022 Nature biotechnology. Single cells are lysed, and RNA is fragmented; Fragments are repaired and polyadenylated, followed by reverse transcription (RT) using barcoded oligo-dT primers; The cDNA is made double stranded and amplified using IVT; aRNA is depleted of rRNA, and libraries are finalized by ligation, RT and PCR, which leave fragments ready for sequencing.