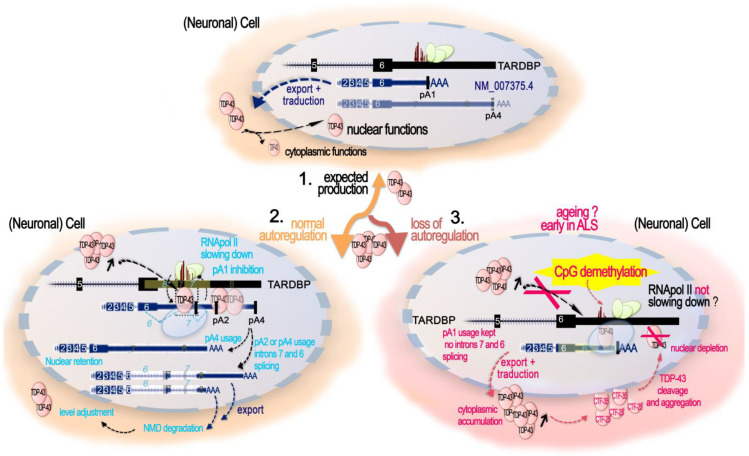
Figure 6.
Role of DNA CpG methylation in TDP-43 levels autoregulation. 1. Expected TDP-43 production. TDP-43 production is promoted by the transcription of an mRNA (blue, NM_007375.4) with 6 exons and usage of poly(A) signal (pA)1 located just after the TDPBR both within the alternative intron 7 (yellow box ≪7≫) or extending up to the pA4. When present at adequate levels, the mRNA is exported to the cytoplasm and is translated to produce TDP-43. 2. Normal autoregulation. When nuclear levels of TDP-43 increase over the expected threshold, TDP-43 binds co-transcriptionally to its own pre-mRNA at TDPBR and down to pA1, stalling RNA pol II at DNA in the correspondence of this region. This impedes pA1 signal usage and facilitates the splicing of the cryptic intron 7. At this stage, three scenario exists: (1) alternative use of pA4 generates an mRNA, mainly retained in the nucleus, with no additional splicing and holding a long 3′UTR; (2,3) pA2 or pA4 are used, and the alternative splicing of cryptic intron 7 and then 6 are triggered. The double-spliced isoforms are exported to the cytoplasm and become degraded via nonsense-mediated mRNA decay. These autoregulatory mechanisms, by resulting in a decrement in the cytoplasmic TDP-43 FL mRNA, allow for the adjusting of the TDP-43 protein levels. 3. Autoregulation failure and role of CpG methylation in the 3′UTR DNA region. The amount of TDP-43 in the nucleus determines the ratio of these isoforms, and when nuclear TDP-43 levels are reduced, the splicing is repressed. The absence of nuclear TDP-43 induces an abnormal autoregulation and increases the amount of TARDBP mRNA in the cytoplasm. The DNA region of the 3′UTR spanning the alternative exon 6-exon 7 junction in correspondence to the TDPBR on pre-mRNA bears a track of 15 CpGs, methylated in the human cortex. When these 15 CpGs are demethylated, TDP-43 autoregulation is negatively affected, suppressing the alternative exon 7 splicing and increasing canonical mRNA levels. The precise mechanisms are not known but possibly involve a reduction in RNA Pol II pausing or alteration in the recruitment of other factors. In vitro, this region is sensible to Tet1-induced demethylation and DNMT3b-induced re-methylation, both of which modulate intron splicing. In the human control motor cortex, DNA methylation at 3′UTR CpGs 10–15 is inversely correlated with age, and, generally, the CNS appears more affected than the liver, showing disparities among regions, with the motor cortex having the lowest CpG 10-15 methylation levels.