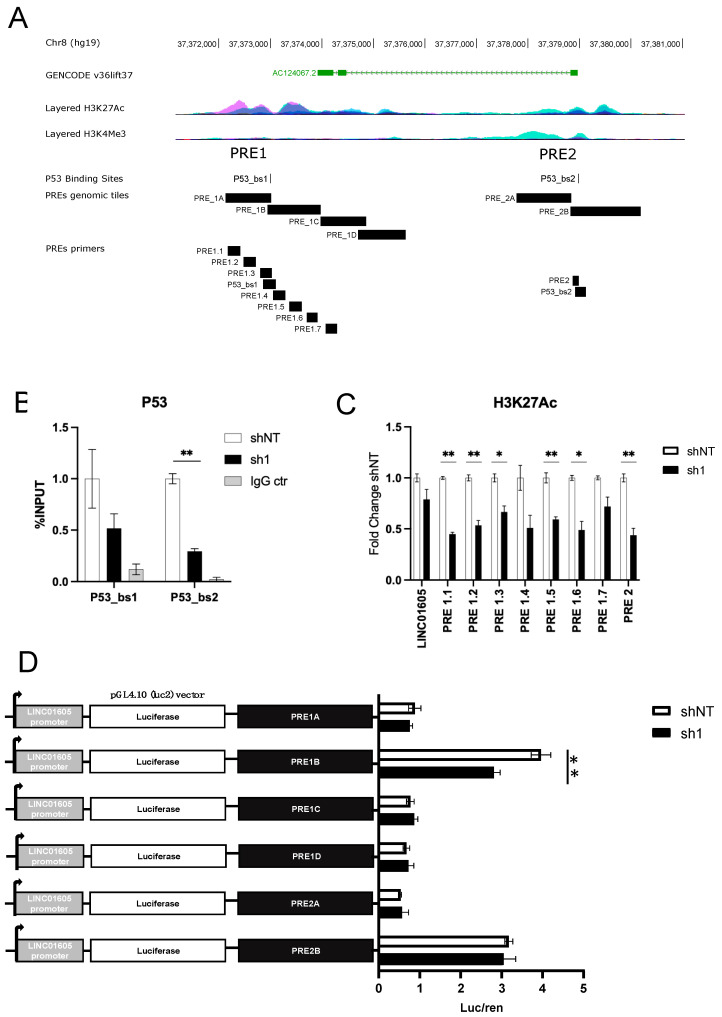
Figure 3.
Identification of PRE1B as mut_p53-dependent enhancer, which regulates LINC01605 expression. (A) UCSC Genome browser session displaying the following from top to bottom: PRE1 and PRE2 overlapping ENCODE H3K27Ac histone mark with putative binding sites for p53 (p53_bs1 and p53_bs2); genomic tiles for PRE1 (PRE1A, PRE1B, PRE1C, PRE1D) and PRE2 (PRE2A and PRE2B) with PRE primers for ChIP and RT-qPCR. (B) ChIP experiment by immunoprecipitating mut_p53 in MDA-MD-231 shNT and sh1 cells and qPCR enrichment at the newly identified mut_p53 binding sites (p53_bs1 and p53_bs2). (C) ChIP H3K27Ac enrichment in MDA-MB-231 shNT and sh1 cells at the LINC016015 locus and at PRE1 and PRE2. Results are the mean of four technical replicates. (D) Luciferase reporter assay results in MDA-MB-231 shNT and sh1 cells for the PREs. Luciferase/renilla values were normalised to the pGl4.10-LINC01605 promoter only construct. Results are the mean of three biological replicates. An empty pGL4.10 (luc2) vector was also transfected into cells to normalize luminescence values (not shown in the figure) (* p-value ≤ 0.05, ** p-value ≤ 0.01).