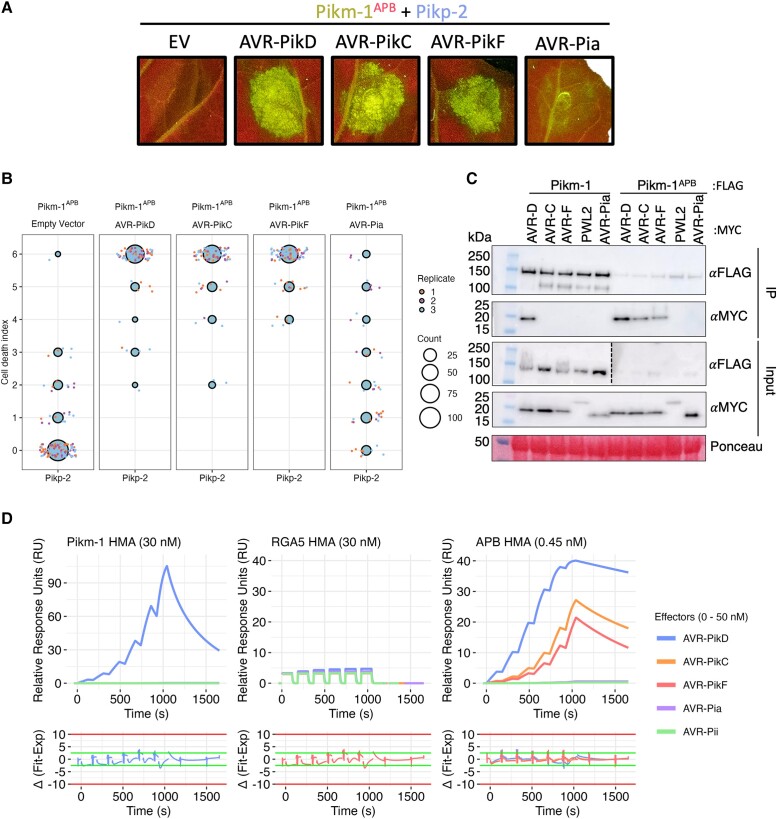
Figure 4.
The RGA5 APB mutant binds and recognizes AVR-Pik when integrated into Pikm-1. A) Pikm-1APB chimera responds to all variants of AVR-Pik tested and activates cell death when coexpressed with Pikp-2 in N. benthamiana, but like Pikm-1RGA5, it only weakly responds to AVR-Pia. B) Cell death scoring of Pikm-1APB coexpressed with AVR-Pik variants D, C, and F in N. benthamiana represented as dot plots. The total number of repeats was 80 per sample. For each sample, all the data points are represented as dots with a distinct color for each of the 3 biological replicates; these dots are jittered around the cell death score for visualization purposes. The size of the central dot at each cell death value is proportional to the number of replicates of the sample with that score. Statistical analyses of these results are shown in Supplemental Appendix S1I. C) co-IP of Pikm-1APB with different MAX (Magnaporthe AVRs and ToxB-like) effectors shows association with AVR-Pik variants, but not AVR-Pia, in planta. Dotted line denotes separate membrane exposures of the same membrane. D) SPR sensograms for the interaction of HMA domains of Pikm-1, RGA5, and RGA5 APB mutant with effectors AVR-PikD, AVR-PikC, AVR-PikF, and AVR-Pia. Non-MAX effector AVR-Pii was added as a negative control. RUs for each labeled protein concentration are shown with the residuals plot beneath (SPR acceptance guides as determined by Biacore software are shown as green and red lines in the residuals plots). Concentration of each protein in the assay is indicated next to their corresponding name. Each experiment was repeated a minimum of 3 times, with similar results.