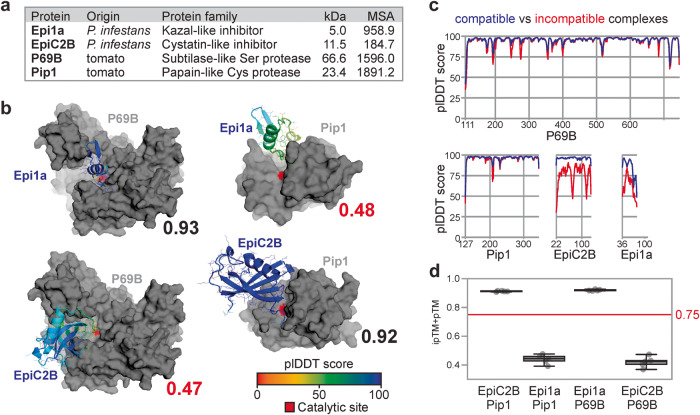
Fig. 1. AFM correctly distinguishes existing from non-existing hydrolase-inhibitor complexes.
a Used inhibitors and their target proteases with their origin, mature molecular weight (MW, in kDa) and depth of mean non-gap multiple sequence alignment (MSA) detected for proteins in compatible complexes. b Best structures predicted by AFM for existing and non-existing inhibitor-hydrolase complexes, with their ipTM + pTM scores ranging from 0 (worst) to 1 (best). Pip1 and P69B are shown in gray, with their catalytically active residue in red. EpiC2B and Epi1a are colored using a rainbow scheme based on their plDDT scores, which range from 0 (worst) to 100 (best). PDB files of these modeals are provided in Supplementary Data 3. c plDDT scores within the four proteins in predicted compatible (blue) and incompatible (red) complexes. d ipTM + pTM quality scores for each of the n = 5 five models for each of the protein pairs, showing the median, 25th and 75th percentiles, and whiskers representing 1.5 times the interquartile range. The raw data are provided in Supplementary Data 6.