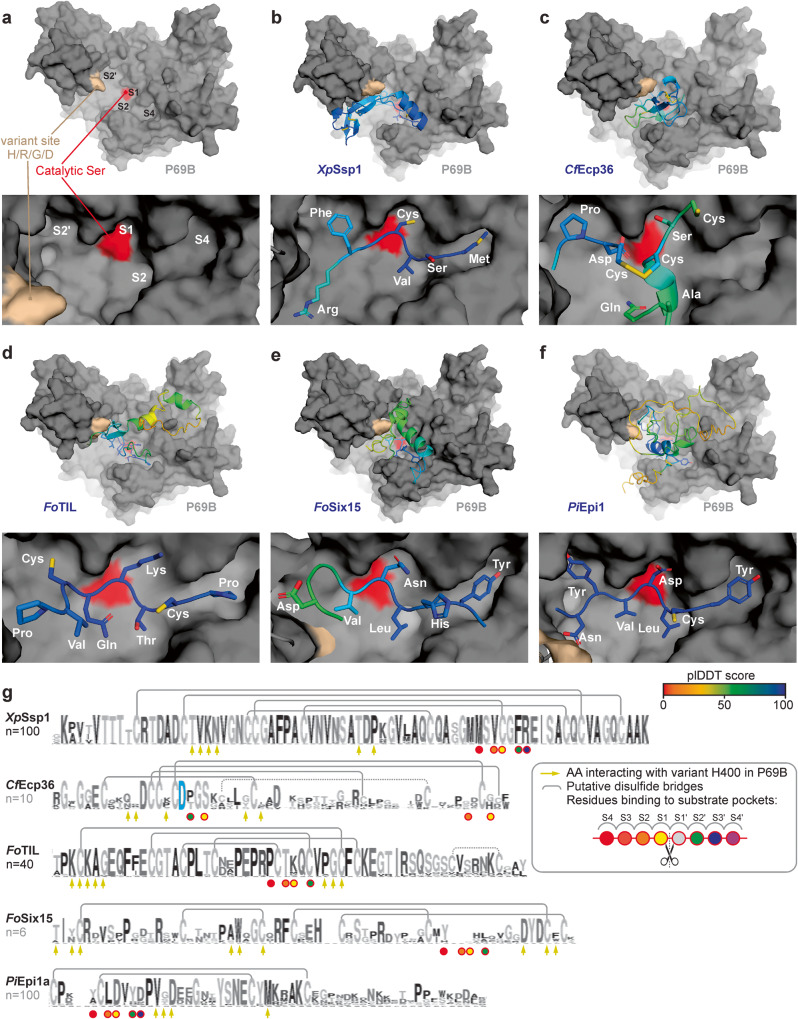
Fig. 5. P69B is an effector hub targeted by five pathogen-derived inhibitors.
AFM-predicted models of P69B without inhibitor (a), or with XpSsp1; (b) CfEcp36; (c) FoTIL; (d) FoSix15 (e) and PiEpi1 (f). P69B is shown in a gray surface representation with the hyper-variant residue (crème) and the active site (red), in the substrate binding groove that has substrate binding pockets (S4-S2-S1-S2’) that bind to substrate/inhibitor residues P4, P2, P1 and P2’, respectively. The inhibitors are shown as cartoons and sticks and colored using a rainbow scheme based on their plDDT scores, which range from 0 (worst) to 100 (best). The zoomed image (bottom) shows the predicted occupation of the substrate binding pockets in P69B by different residues of the inhibitor. g Sequence conservation between homologs of the identified P69B inhibitors. Shown is the sequence logo for n = x close homologs from other plant pathogen species, identified by BLAST searches in the NCBI database and presented in Weblogo95. Highlighted are the residues that probably interact with the substrate binding pockets in P69B (circles); the conserved Asp residue in CfEcp36 that interacts with the catalytic site (blue); the residues that might interact with the variant residue in P69B (arrows); and putative disulfide brides observed in the AFM model (gray lines). PDB files for the shown models are available in Supplementary Data 3.