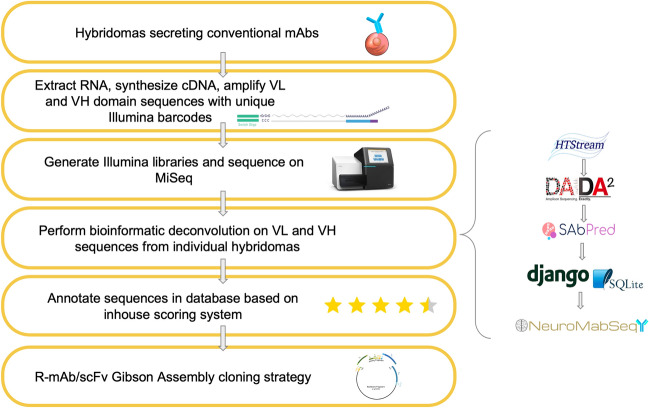
Figure 1.
Sequencing workflow and bioinformatics processing. Hybridomas of interest are sequenced using a workflow consisting of RNA extraction, cDNA synthesis, and semi-nested PCR amplification with IgG-specific primers followed by the addition of unique Illumina barcodes to each sample. Illumina libraries are then generated, and adapters are ligated for sequencing on the MiSeq platform. Bioinformatics processing is shown on the right panel. Reads from the Illumina sequencing are run through HTStream for base quality trimming and other read processing. Next, they are passed through DADA2 for amplicon denoising followed by SAbPred ANARCI tool based on the IMGT numbering scheme. All ASVs, metadata, and other quality metrics are uploaded to the NeuroMabSeq database and website where further information and tools are provided to the end users. This includes but is not limited to BlastIR results, BLAT searches across the database, and recommended high quality sequences for recombinant antibody design. Annotations of internally generated scores are provided in addition to other database information. Finally, high quality sequences are used in the design of gene fragments for generation of R-mAb and scFv expression plasmids.