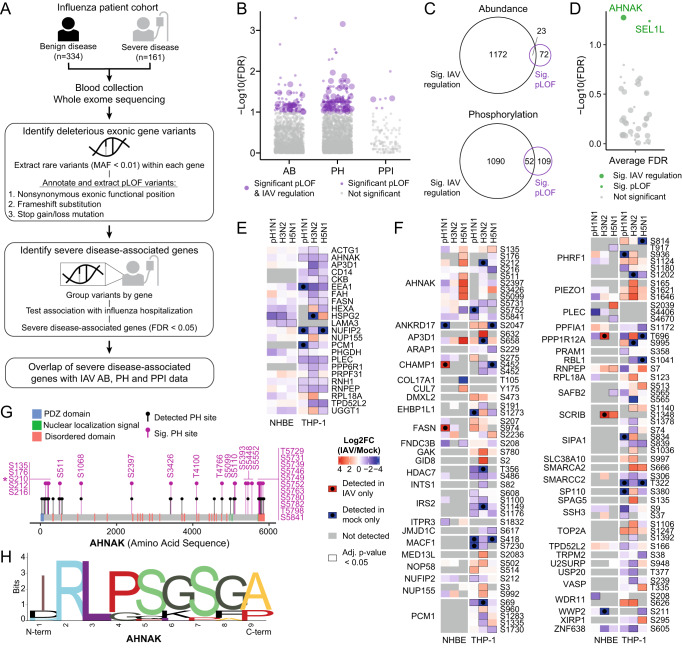
Fig. 4. Patient exome sequencing identifies gene variants encoding proteins that are regulated in AB and PH during IAV infection.
A Schematic representation of sample collection and data analysis for identifying genes with pLOF variants associated with severe influenza disease from an influenza patient cohort. Genes with pLOF variants plotted against the false discovery rate (-log10(FDR)) from the severe-disease association test for (B) each of the AB, PH and PPI datasets or (D) the average FDR across the three AB, PH, and PPI datasets (purple and green circles=genes with significant pLOF variants (FDR < 0.1); grey=genes with pLOF variants below threshold (FDR > 0.1); large circles=genes with significant protein AB or PH changes (adjusted p-value < 0.05; two-sided t-test); small circles=genes detected in AB or PH proteomic datasets with no significant changes). C Venn diagram of the overlap of proteomic datasets with significant changes in AB (top, left) or PH (bottom, left), and significant genes with pLOF variants (corresponds to the total number of purple circles in B). Heatmap of log2FC in infection vs mock (log2FC (IAV/Mock)) from NHBE and THP-1 cells at 18 hours (pH1N1, H3N2) and 12 hours (H5N1) post-IAV infection (reported in Supplementary Data 2) for (E) AB of 23 significantly changing proteins, and (F) PH of 52 changing phosphorylated proteins, that have significant pLOF variants (from the union in C) (red=increase, blue=decrease, grey=not detected; red box with black circle=only detected in IAV infection, blue box with black circle=only detected in mock infection; black box outline=significant change (adjusted p-value < 0.05; two-sided t-test)). G AHNAK phosphorylation sites detected and significantly changed in the PH data (black pins=detected no significant change; pink pins=significantly changed (adjusted p-value < 0.05; two-sided t-test); asterisk=AHNAK S210). H Multiple sequence alignment sequence LOGO (S210P/Q, middle) for phosphorylation disruption mutations in AHNAK created with WebLogo version 2.8.2148 using motifs identified by pLOF analysis as likely to be loss of phosphorylation.