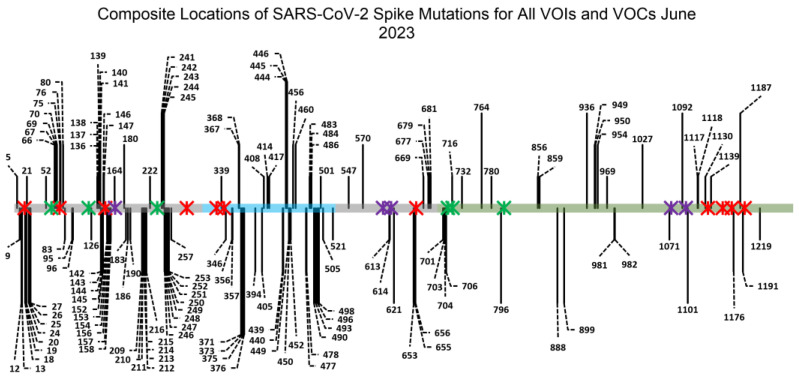
Figure 1.
Representation of the SARS-CoV-2 spike gene amino acid sequence with all locations of VOC mutations. The S1 domain is depicted in grey (residues 14 to 685), which is further subdivided into four domains: the N-terminal domain (NTD, residues 14 to 306), the receptor-binding domain (RBD, residues 319 to 527, depicted in teal) and the two C-terminal domains, C-terminal domain 1 (CTD1, residues 527 to 591) and C-terminal domain 2 (CTD2, residues 591 to 686). Within the receptor-binding domain is the receptor-binding motif (RBM, residues 438 to 506), which encompasses the key amino acids that mediate SARS-CoV-2 spike protein binding to the host hACE2 receptor. The S2 domain is depicted in grey-green and ranges from residues 686 to 1273. Black lines indicate the locations of point mutations across VOCs (for a list of the specific VOCs summarized here, see Figure S2). Red, green and purple Ж crosses depict the locations of N-linked glycans. Red crosses indicate complex glycans, green crosses indicate high-mannose glycans and purple crosses depict hybrid glycans and glycans that were inconsistently identified as being complex or high-mannose, based on an analysis of existing literature (Figures S3 and S4). While mutations have occurred in VOCs immediately adjacent to glycan sites, there have been no variants that have mutations at the asparagine residues where N-linked glycosylation is present [41,55,56].