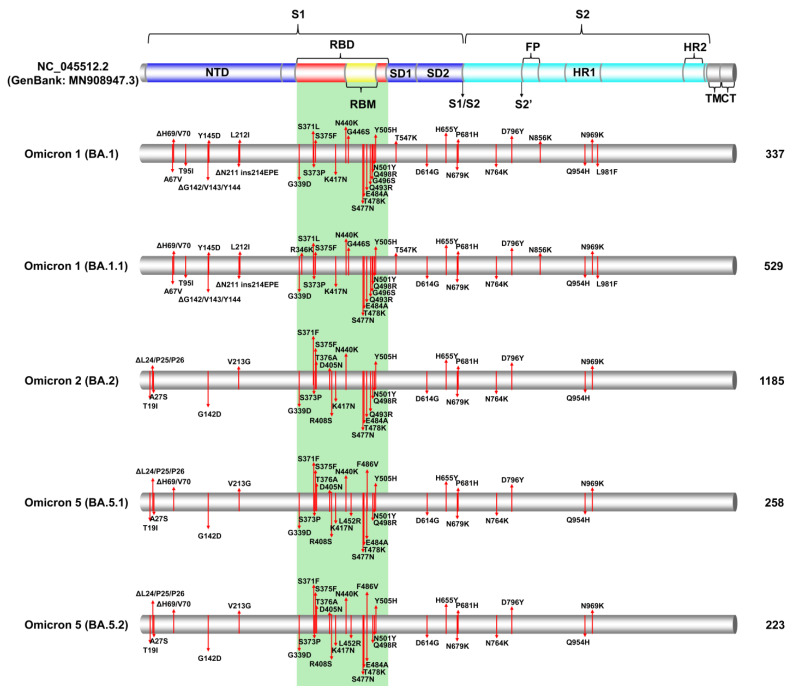
Figure 3.
The figure effectively illustrates the significant S protein mutations within these prominent SARS-CoV-2 lineages during the specified period, shedding light on the molecular characteristics of the virus during the 5th and 6th waves in Cyprus. These mutations were identified through the analysis of Cypriot SARS-CoV-2 sequences collected between October 2021 and October 2022. Specifically, the figure focuses on the most common S protein mutations found in Omicron 1 (BA.1, BA.1.1), Omicron 2 (BA.2), and Omicron 5 (BA.5.1, BA.5.2) lineages. A colored cylinder represents the essential domains of the SARS-CoV-2 S protein (GenBank: MN908947.3), including the N-terminal domain (NTD); the receptor-binding domain (RBD), highlighted in red; the receptor-binding motif (RBM) in yellow; subdomains 1 and 2 (SD1 and SD2); fusion peptide (FP); subunit 1 (S1) in blue; subunit 2 (S2) in cyan; heptad repeats (HRs); the transmembrane domain (TM) in gray; and the cytoplasmic tail (CT) in gray. Cleavage sites (S1/S2 and S2′) are marked with black arrows. The green-highlighted area corresponds to the receptor-binding domain (RBD) [42,43,44,45,46,47,48,49,50,51]. Mutations are indicated by red lines, denoting the locations of the most common mutations identified in all or nearly all sequences within a given lineage. The figure also provides information on the total number of sequences analyzed for each lineage in this study, displayed on the right side of the figure.