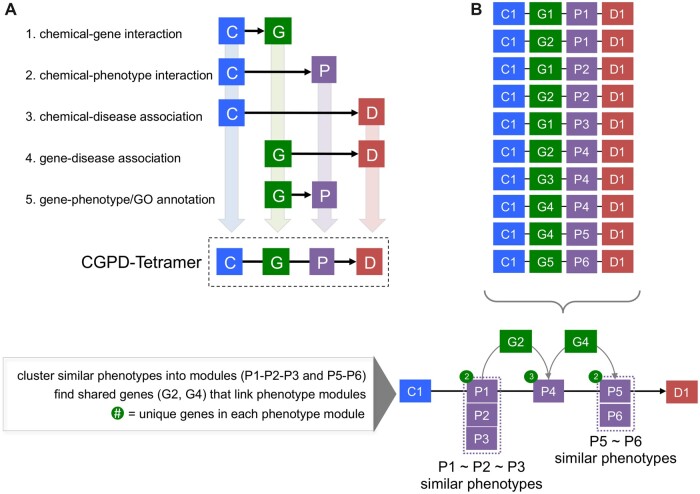
Figure 1.
CGPD-tetramers. A, CGPD-tetramers are computationally generated information units that interrelate 4 data types from CTD: a chemical (C), gene product (G), phenotype (P), and disease (D). To generate a CGPD-tetramer by data integration, 5 individual lines of supporting evidence are required as directly curated interactions between the 4 data types: chemical-gene, chemical-phenotype, chemical-disease, gene-disease, and gene-phenotype/GO. If any 1 line of supporting evidence is lacking, the tetramer is not generated. B, Maximizing the shared gene and phenotype overlaps between individual tetramers allows them to be constructed into longer, more detailed molecular mechanistic pathways (see Davis et al. [2020] for details). Here, 10 tetramers involving 1 chemical (C1), 5 unique genes (G1–G5), 6 phenotypes (P1–P6), and 1 disease outcome (D1) can be combined. Similar phenotypes (eg, P1–P2–P3 and P5–P6) can be clustered into separate modules. Shared genes are used to bridge the modules through shared molecular mechanisms: gene G2 connects phenotype module P1–P2–P3 to phenotype P4, and then gene G4 bridges phenotype P4 with phenotype module P5–P6. Some examples of similar phenotypes might include all those involving cell death, such as “apoptotic process” (GO:0006915), “apoptotic signaling pathway” (GO:0097190), “programmed cell death” (GO:0012501), “positive regulation of apoptotic process” (GO:0043065), “cell death” (GO:0008219), etc.