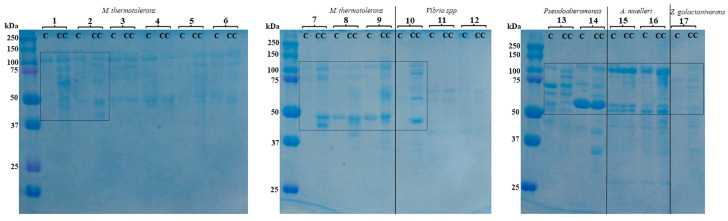
Figure 3.
SDS–PAGE analysis of the culture supernatants: lane kDa, protein size markers (BioRad); lanes C—control culture supernatants grown without colloidal chitin, lanes CC—culture supernatants grown in the presence of colloidal chitin. The lanes under square brackets 1—Microbulbifer thermotolerans Sgf 25; square brackets 2–6—M. thermotolerans Sh 1–5; square brackets 7—M. thermotolerans JCM 14709T; square brackets 8—M. thermotolerans Sgm 25/1; square brackets 9—M. thermotolerans 14G-22; square brackets 10—Vibrio sp. Sgm 5; square brackets 11—Vibrio sp. 14G-20; square brackets 12—Vibrio sp. Sgm 22; square brackets 13—Pseudoalteromonas sp. B530; square brackets 14—Pseudoalteromonas flavipulchra KMM 3630T; square brackets 15—Aquimarina muelleri V1SW 51; square brackets 16—A. muelleri V1SW 49T; square brackets 17—Zobellia galactanivorans DjiT. Predicted induced chitin-degrading enzymes compared to control are underlined by squares with broken line. Protein was concentrated to 30 µg with 10% trichloroacetic acid and applied into the well.