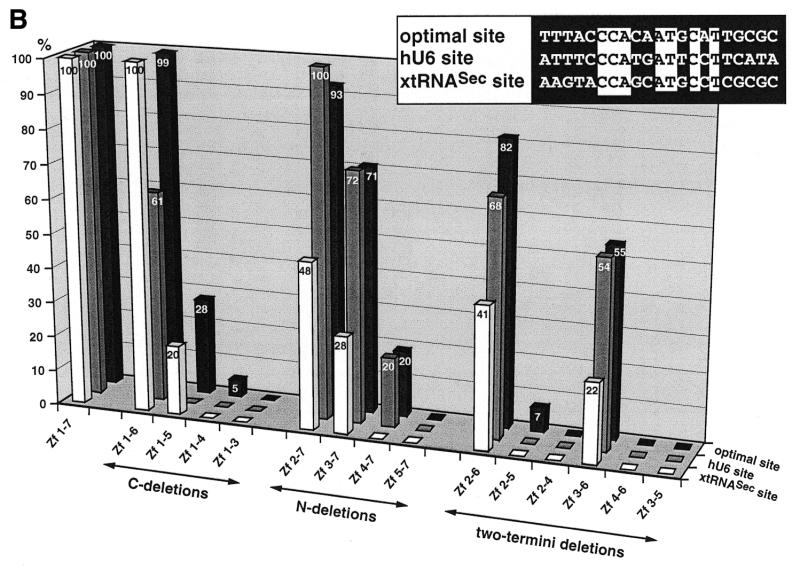
Figure 1.
Binding of wild-type and truncated Staf zinc finger domains to the optimal Staf-binding site and to the Staf motifs in the human snRNA U6 (hU6 site) and in the X.laevis tRNASec (xtRNASec site) promoters. (A) Amino acid sequence of residues 255–476 displaying the sequence alignments of the seven zinc fingers (18). Gaps (<) have been introduced at two locations to maximize the match. Cysteines, histidines and invariant hydrophobic residues are depicted in bold. Open and solid triangles indicate the N- and the C-termini of the various Staf zinc finger domains expressed as GST fusion proteins in E.coli, respectively. (B) Relative binding efficiencies of wild-type and truncated Staf zinc finger domains for the optimal, hU6 and xtRNASec sites. The histogram plots the amount of probe bound to the truncated proteins, relative to that obtained with the wild-type zinc finger domain Zf 1–7. Binding reactions contained 2 nM DNA and 60 nM fusion protein. The results of one representative experiment for each protein and probe are shown. Two other independent determinations gave similar results. Sequence comparisons between the optimal, hU6 and xtRNASec Staf-binding sites used in this study are shown in the upper right pannel. Nucleotide identities between the different elements are boxed.