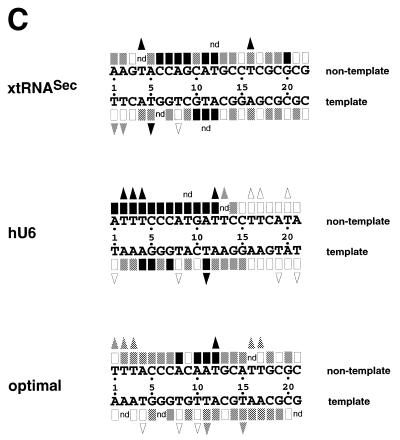
Figure 2.
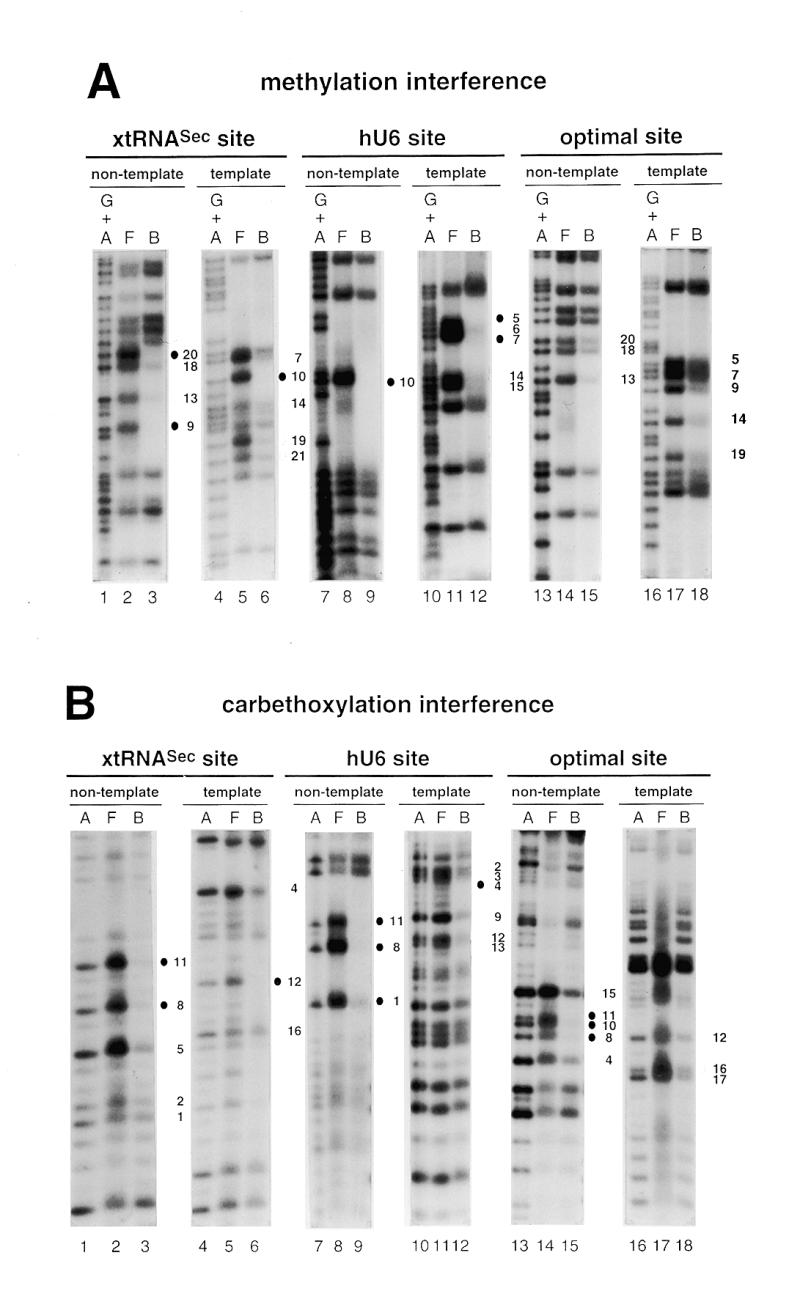
Interference with Staf binding by methylation, carbethoxylation, KMnO4 modification and depyrimidation of DNA fragments containing the xtRNASec, hU6 or optimal Staf-binding sites. The DNA fragments were 32P-labeled at their 5′-ends on the template or non-template strands, partially methylated (A) or carbethoxylated (B) and subjected to interference analysis. DMS modification and treatment of the modified DNA were performed under conditions allowing cleavage at only the modified guanine bases. Lanes A and G + A, sequencing reactions; lanes F and B, free and bound DNAs. Bases whose modifications interfered with Staf binding are shown by dark (total interference) or gray (partial interference) circles. Bases are numbered according to the Staf consensus binding site (21). (C) Summary of the interference experiments. The full and partial interference effects of guanine methylation, adenine carbethoxylation and pyrimidine removal on Staf binding are represented by dark and gray squares, respectively. The full and partial interference effects on the binding of Staf by KMnO4 modification of thymines are depicted by dark and gray triangles, respectively. Open squares and triangles show positions where base modifications or removal did not interfere with Staf binding; nd, not determined.