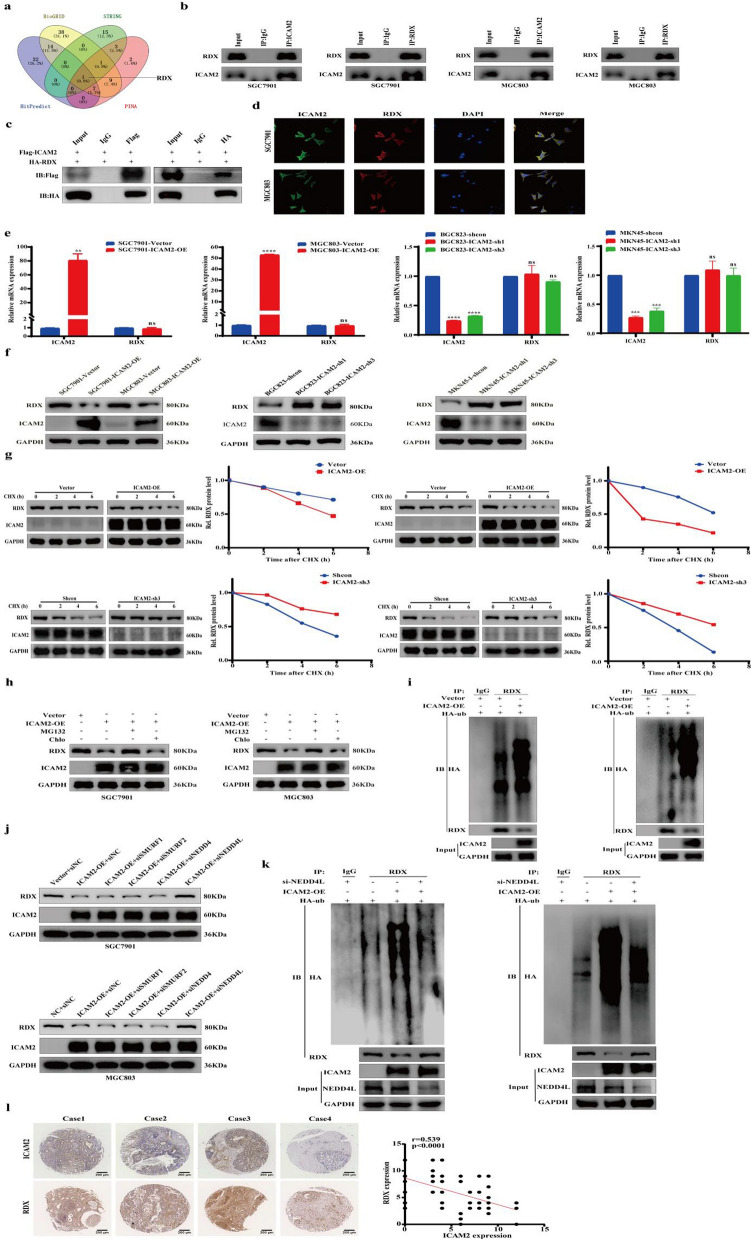
Fig. 5.
ICAM2 mediates RDX ubiquitination degradation in a NEDD4L-dependent manner. a Bioinformatic analysis of potential interacting proteins of ICAM2. b Co-IP was conducted to determine the interaction between endogenous ICAM2 and RDX in SGC7901 and MGC803 cells. c Co-IP was performed to detect the interaction between exogenous ICAM2 and RDX by co-transfecting Flag-ICAM2 and HA-RDX plasmids into 293 T cells. d Immunofluorescence staining examining the colocalization of ICAM2 and RDX in GC cells. e Quantification of RDX mRNA expression using RT-PCR following ICAM2 overexpression or knockdown. f Western blot analysis of RDX protein expression after ICAM2 overexpression or knockdown. g CHX chase assay to explore the half-life of RDX protein in the presence or absence of ICAM2. h Evaluation of the stability of RDX protein through chloroquine or MG132 treatment in the control and ICAM2 overexpression groups. i RDX ubiquitination level was detected by the Co-IP and Western blotting assays in the ICAM2 overexpression and control groups. j Western blot analysis investigating the effects of knockdown of the main E3 ubiquitin ligases on RDX protein levels in ICAM2 overexpression GC cells. k Assessment of the ubiquitination level of RDX in GC cells with ICAM2 overexpression following NEDD4L knockdown. l Representative images of ICAM2 and RDX immunohistochemically stained in paired GC tissues. Linear regression analysis of ICAM2 and RDX expression