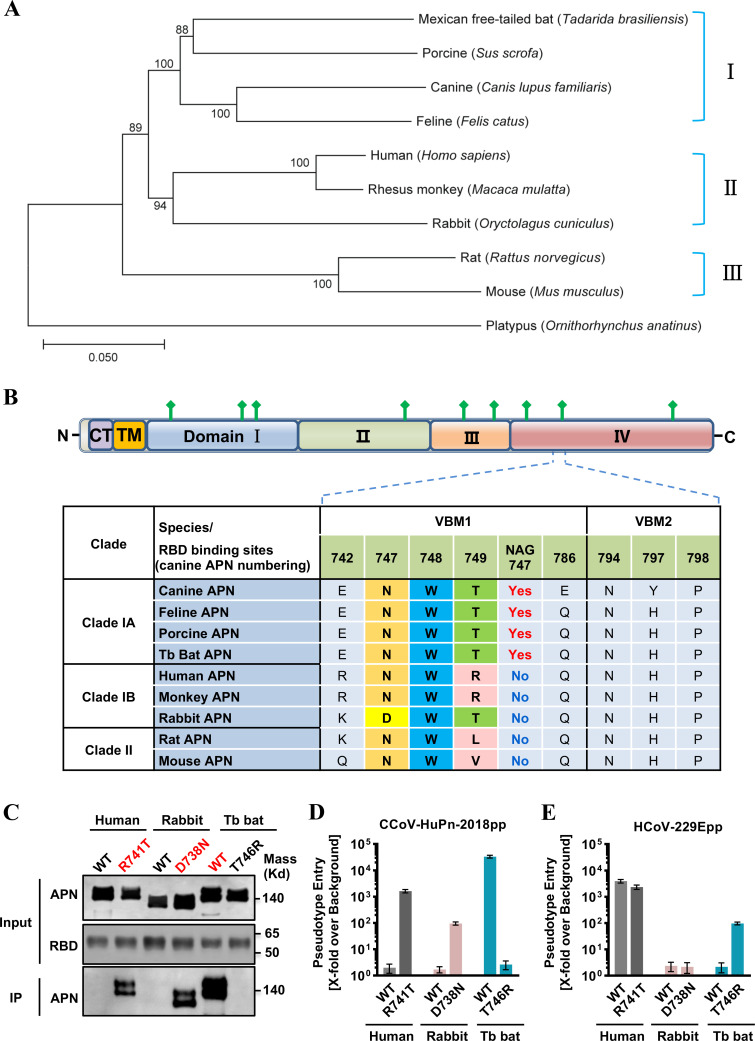
FIG 3.
Glycosylation at residue 747 of APN is critical for its receptor activity. (A) Phylogenetic analysis of APN orthologs. Phylogenetic tree was constructed based on nucleotide sequences of the complete APN gene using the neighbor-joining method implemented in MEGA7.0.26 software. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree was rooted by the APN of a platypus (Ornithorhynchus anatinus, GenBank No. XM_029051618.1). The animal species are shown on the right-hand side of the tree. (B) At the top is the domain organization of the canine APN. APN is a type II membrane glycoprotein that has a short N-terminal intracellular tail, a transmembrane domain, and a large C-terminal ectodomain. CT, cytoplasmic tail; TM, transmembrane domain; domains I, II, III, and IV are in blue, green, light orange, and light red, respectively. N-linked glycans are rendered as green protrusions. At the bottom is the sequence alignment of nine APNs focused on two VBMs and N747 glycan (canine APN numbering). One-letter codes are used for amino acids, and amino acids of the “N-W-T” motif are highlighted with different colors. (C) IP assay. The upper panel shows the input of wild-type APN proteins and APN mutants with a C9 tag and RBD with an IgG Fc tag (RBD-Ig). The lower panel shows the APN pulled down by the RBD-Ig fusion protein. (D and E) VSV-Luc-based pseudotyped virus entry. 293T cells were transfected with wild-type APN orthologs (human APN, rabbit APN, and bat APN) or APN mutants (human APN R741T glycan knockin mutant, rabbit APN D738N glycan knockin mutant, and bat APN T746R glycan knockout mutant). At 48 h post-transfection, the cells were infected by CCoV-HuPn-2018pp (D) or HCoV-229Epp (E). At 24 h post-infection, luciferase signals were measured, and pseudotype entry was normalized against the background (signal obtained for pseudotyped particles without viral envelope protein). Error bars reveal the standard deviation of the means from three biological repeats.