Figure 1.
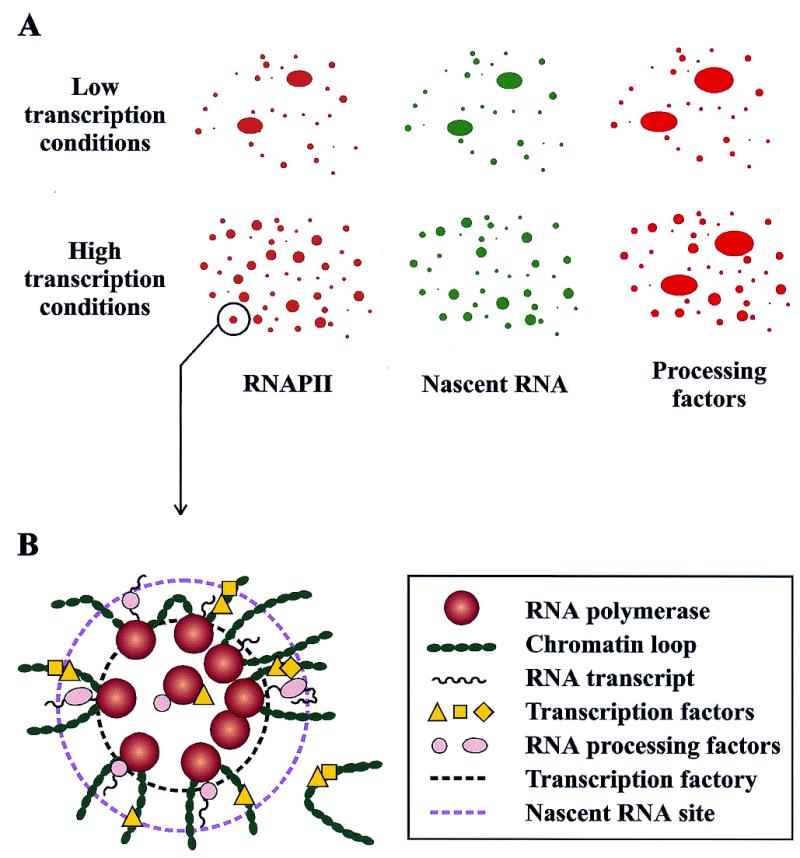
Schematic representation of the subnuclear localization of pre-mRNA transcription and processing components as observed by immunofluorescence labeling and confocal microscopy analyses. (A) Immunodetection with antibodies to the RNAPII phosphorylated CTD reveals RNAPII localized in the nucleus in thousands of tiny clusters. These foci, which occupy <1% of the nuclear volume, overlap with the sites of pre-mRNA synthesis detected by labeling the nascent transcripts with BrdU. In general, transcription and processing factors colocalize with the RNAPII foci, although particular factors can be observed at other locations that may represent storage pools. In cells that are actively transcribing, RNAPII transcription foci are diffusely distributed throughout the nucleus (bottom). When transcription is inhibited, sites contract to minimal domains (top). (B) Two-dimensional schematic representation of an RNAPII transcription factory (inner dashed circle) and its surrounding region of chromosome loops and transcripts with associated processing factors (outer dashed circle). In this model, genes are recruited to and transcribed by attached polymerases. Most genes are transcribed by a single RNAPII, although particular genes may have several transcribing polymerases.
