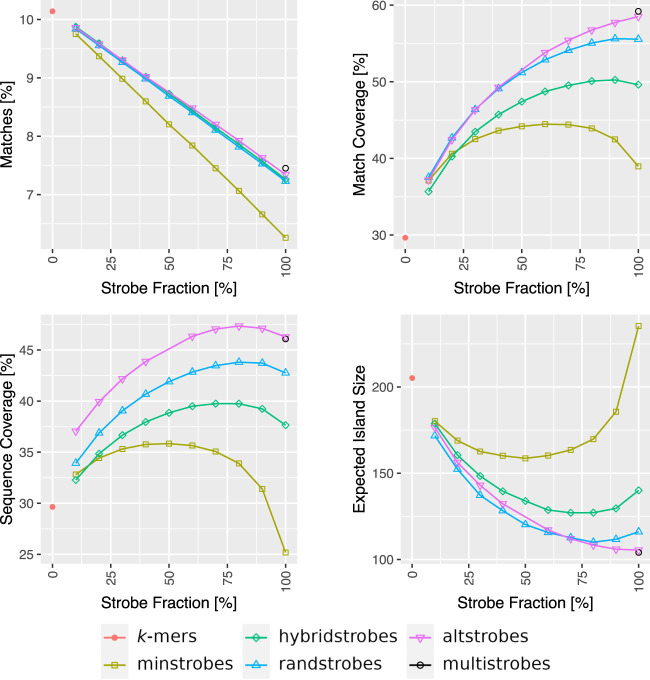
Figure 4.
Comparison between (mixed-)strobemers (2,15,25,50, q), (mixed-)altstrobes (2,10,20,25,50, q), multistrobes (2,5,25,25,50), and k-mers (k = 30) when mapping genomic Oxford Nanopore Technology (ONT) reads from E. coli to its reference. The E. coli reads were split up in long disjoint segments of 2000 nt. Next, the segments were seeded with strobemer fractions q from 0% (k-mers) to 100% (strobemers), downstream windows set to [25,50] and all strobes combined adding up to equal length subsequences of size 30 for better comparison. Then for each segment, the collinear solution of raw hits was computed to subsequently quantify the number of matches, match coverage, sequence coverage, and expected island size.