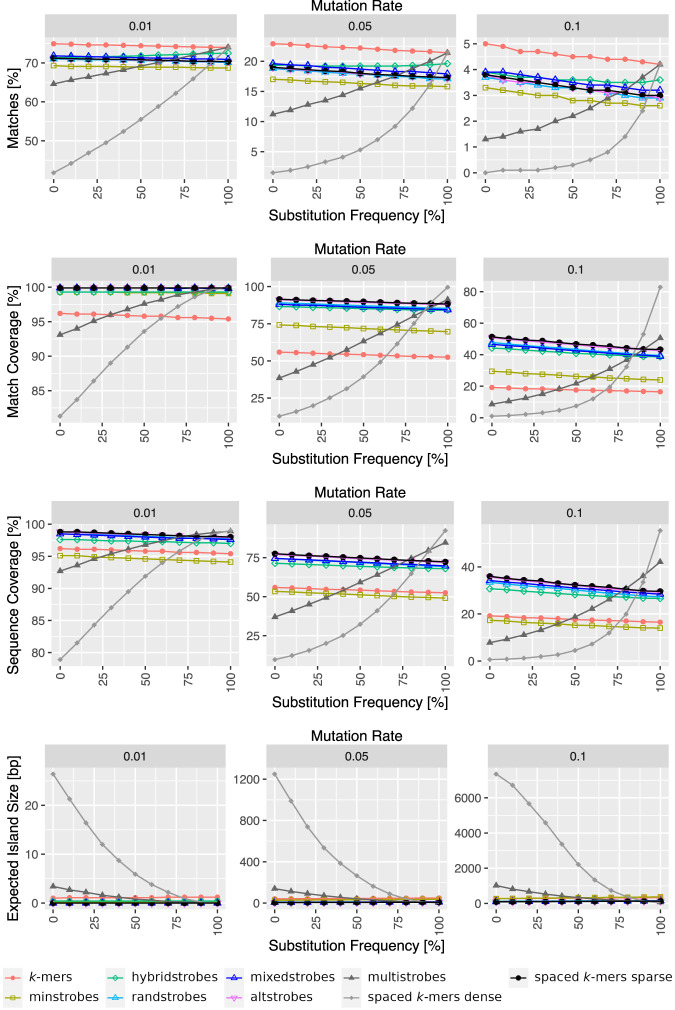
Figure 5.
Performance of (spaced) k-mers and strobemers with various indel/substitution patterns in simulated data. For all the experiments, 1000 random sequences of length 10,000 nt were created and 1%, 5%, and 10% of the nucleotides of the reference string were mutated for the different experimental conditions. Substitutions were hereby added with probabilities from 0% to 100% (substitution frequency), whereas the other mutation positions were filled with insertions and deletions with equal probability. Subsequently, the sequences were seeded with (spaced) k-mers of length k = 30 as well as strobemers of order n = 2 with all sub-strobes summing up to k = 30 and downstream windows set to [25,50].