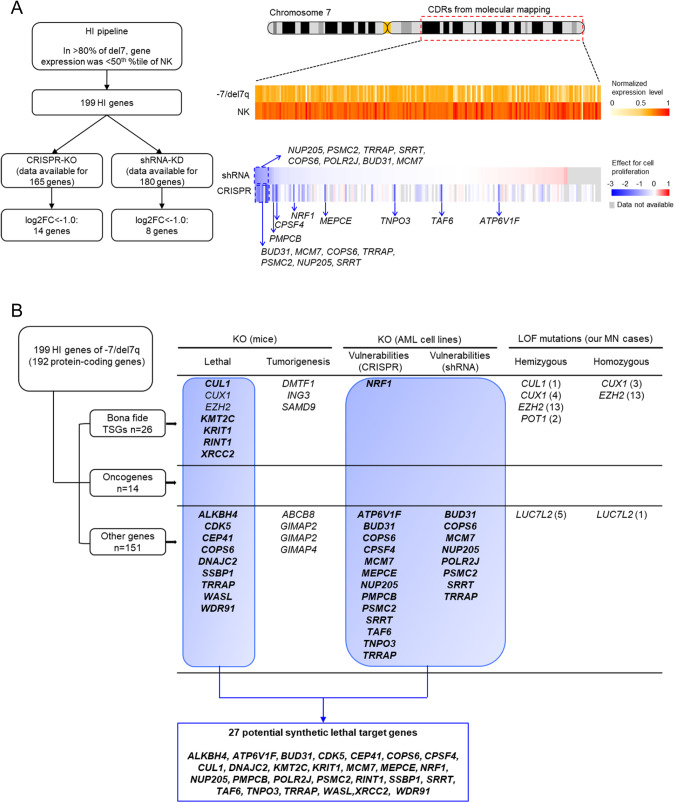
Fig. 7. Multiple haploinsufficient (HI) genes involved in leukemia cell survival and potential synthetic lethal target genes for -7/del7q myeloid neoplasm.
A Flowchart (left) for selecting the genes, CRISPR/shRNA knockout (KO) of which showed significant cell growth inhibition or a trend towards cell proliferation. Graphic (right) depicting the differentially gene expression levels between -7/del7q and NK myeloid neoplasms of the 199 HI genes in the CDRs identified by molecular mapping and the effect of shRNA or CRISPR KO of the 199 HI genes on cell proliferation from DepMap public database (https://depmap.org/portal/). B Summary of 199 HI genes based on lethality in KO mice collected from literature review, vulnerability of leukemia cells by CRISPR/shRNA KO screens from DepMap database, and the presence of cases affected by hemizygous or homozygous LOF mutations in our cohort. The numbers in brackets indicate the number of cases with each hemizygous or homozygous mutation in our cohort.