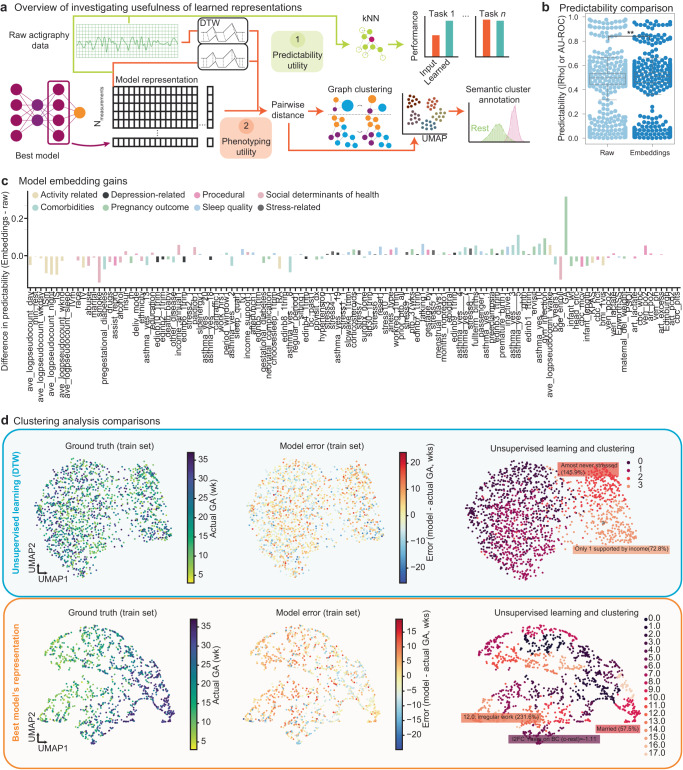
Fig. 4. series2signal module to investigate model utility demonstrates that series2signal embeddings are useful for predicting ancillary metadata tasks and for phenotyping patients.
a Overview of model utility module that compares an unsupervised learning approach harnessing dynamic time warping (DTW) to measure distances between pairs of time-series with supervised learning and graph clustering approach based on representations of input actigraphy data that our model learns. b Predictability, defined as AU-ROC for classification tasks and the absolute value of Spearman’s ρ for regression tasks, of metadata tasks using input, pre-processed actigraphy data (Raw) or learned model embeddings (Embeds). Box plots show median and first and third quartiles with outliers as 1.5 times IQR. c The difference in predictability between models built using learned model representations (Embeddings) minus those built with input actigraphy. Color shows category of metadata target. d Dimensionality reduction of actigraphy data using DTW (top row) versus model embeddings (bottom row) with individual measurement-GA pairs in the train set colored by actual GA, error, and graph-based clustering with semantic identification of cluster enrichment across metadata variables (columns). Far right numbers indicate cluster ID.