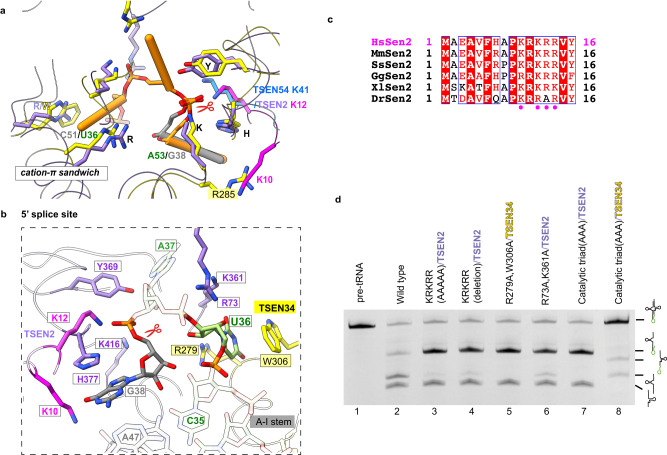
Fig. 5. Recognition and cleavage mechanisms of 5′ splicing boundary by human TSEN complex.
a Overlay of the catalytic centers of TSEN34 and TSEN2. The 3′ bugle in TSEN34 is shown in orange, while the nucleotide G38 in TSEN2 is in gray. TSEN2 was used for the overlay. Key residues that are involved in recognition and cleavage are shown. K10 and K12 in the KRKRR motif of TSEN2 play similar roles for the 5′ splice site as R285 of TSEN34 and K41 of TSEN54 for the 3′ splice site. b Detail view of the 5′ splice site with TSEN34 and TSEN2. U36 interacts with TSEN34 R279 and W306 via cation-π interactions. The scissile phosphate is located in the center of the catalytic triad of TSEN2 –Y369, H377, and K416. A37 that is not visible in the density map is shown as transparent sticks. c Sequence alignment of the N-terminal insertion of selected vertebrate TSEN2 homologs. Well-conserved residues are highlighted in red. The positively charged motif, close to the intron loop, is indicated in magenta dots. Hs: human; Mm: mouse; Ss: pig; Gg: chicken; Xl: frog; Dr: zebrafish. d Cleavage assays were carried out to assess the impact of various mutations on the cleavage activity at the 5′ splice site. The mutation of the catalytic triad of residues in TSEN2 (lane 7) resulted in the complete loss of cleavage activity. Deleting the KRKRR motif of TSEN2 or mutating it to AAAAA greatly reduced the cleavage activity at the 5′ splice site (lanes 3 and 4). The mutation of the R279-W306 tweezer in TSEN34 led to the complete abolishment of cleavage at the 5′ splice site (lane 5). Mutating R73A and K361A in TSEN2 slowed down the cleavage rate (lane 6). For reference, lane 8 represents the result obtained from mutating the catalytic triad of residues in TSEN34. These cleavage assays were repeated three times. The reactions were carried out at 37 °C for 10 min and the molar ratio of RNA to TESN was 1:1. Source data are provided as a Source data file.