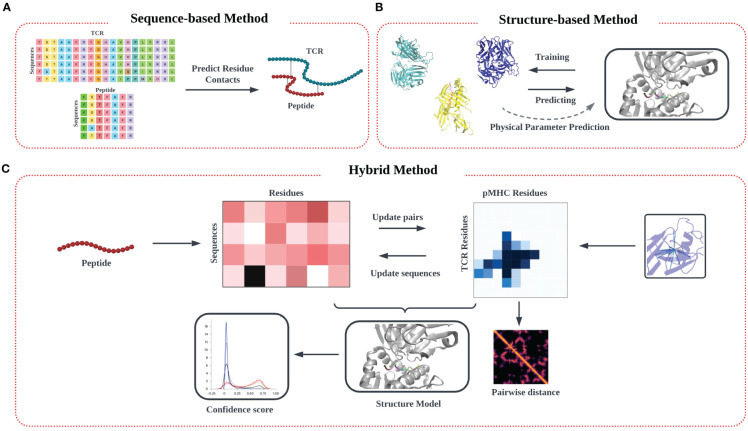
Figure 1.
Modeling approach to TCR-pMHC prediction based on input data. (A) Models trained purely on TCR and peptide sequence input data feature multiple sequence alignment (MSA) on input data matrices to identify patterns, followed by identification of potential interaction pairs using various algorithms techniques. (B) Models trained on input structural data models commonly aim to identify the TCR-pMHC binding interface, along with associated information on binding affinity. Predictions are often made by determining similarity in secondary structure in the interfacial region of the binding interface, from which physical parameters like binding affinity and kinetic data (KD , Koff ) can be estimated. (C) A third ‘hybrid’ category of inferential model synergistically combines sequence and structural data in the training step.