Fig. 2 -. Deployment of a genetically encoded heme biosensor in Cryptococcus neoformans.
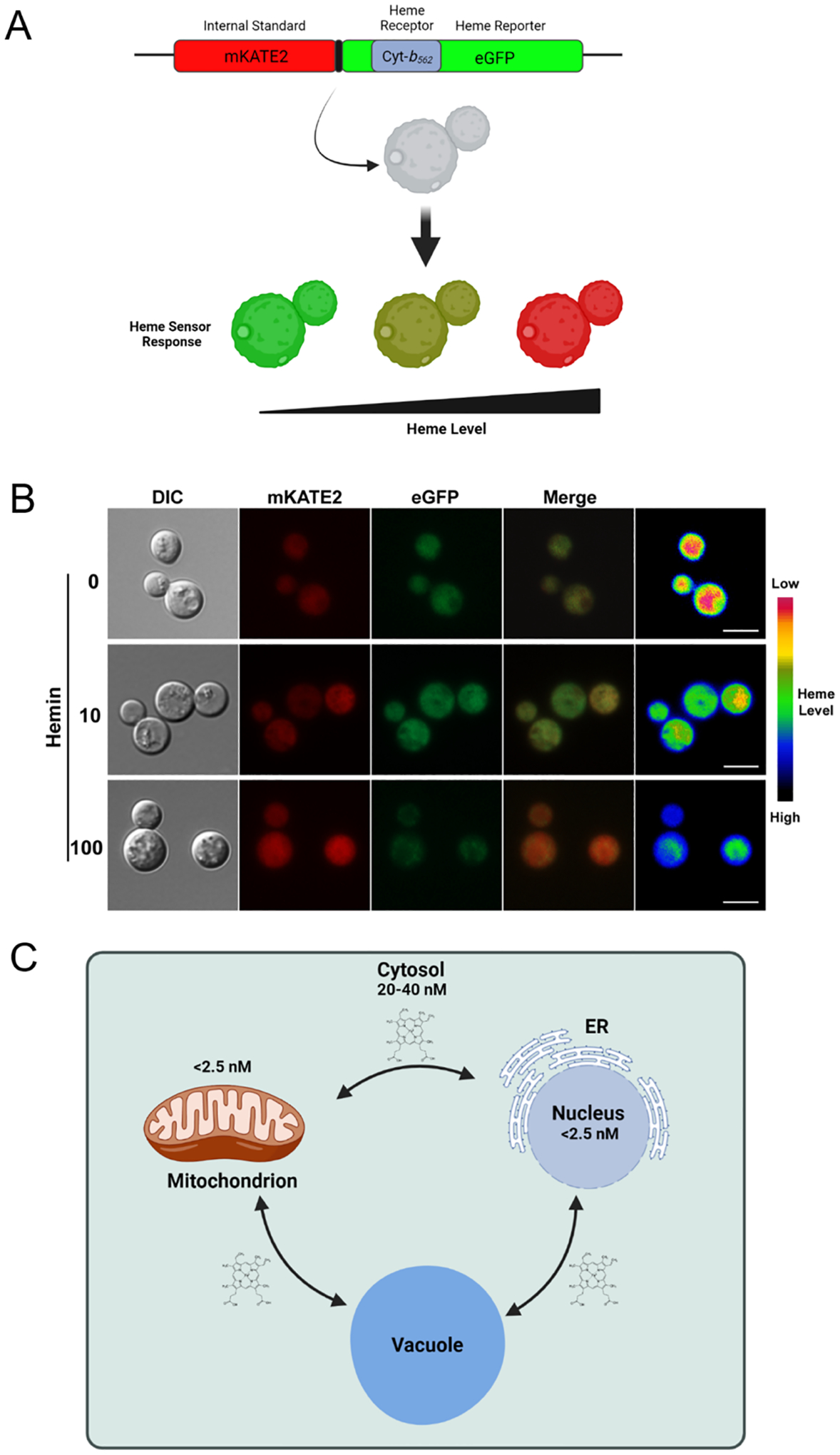
(A) Diagram of sensor composition including the heme binding domain of Cyt-b562 within eGFP and the mKATE2 protein to allow ratiometric measurements (Hanna et al., 2016). Heme binding quenches the GFP signal but does not influence mKATE2 thus resulting in a shift in the ratio of fluorescence from the two proteins. (B) Example of the evaluation of the labile heme pool in C. neoformans by microscopy. The images are from a replicate of an experiment presented by Bairwa et al. (2020). Scale bar = 5 μm. (C) Diagram of cellular compartments and labile heme concentrations estimated by Hanna et al. (2016) for S. cerevisiae.
