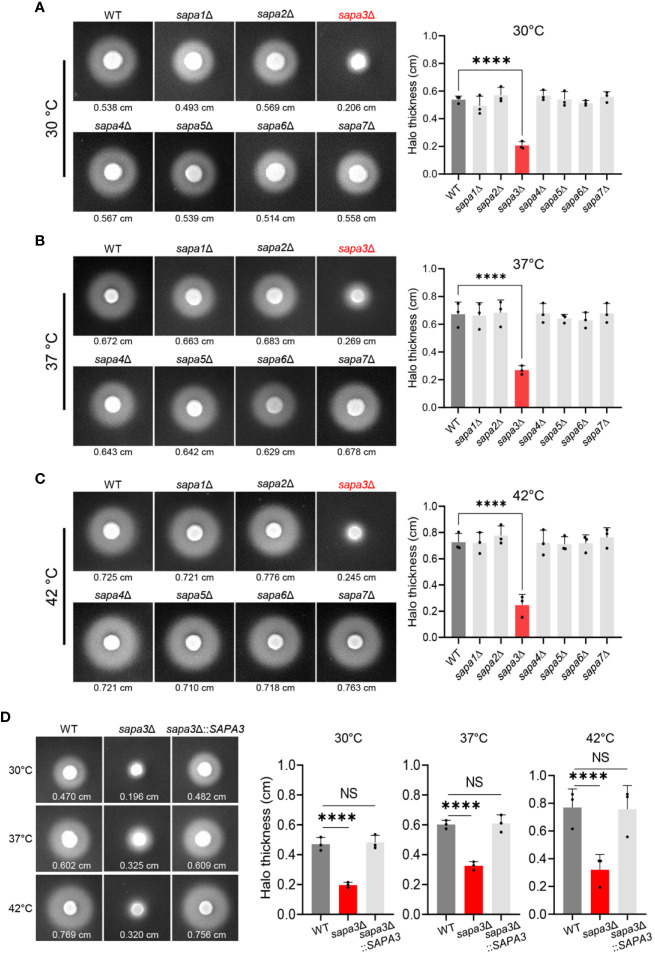
Figure 3.
Role of Sapa3 as a key secreted aspartyl proteinase in C. auris. (A–C) Comparative analysis of SAP activities in wild-type and mutant strains of C. auris at different temperatures. Overnight cultured cells of the wild-type strain and SAP gene mutants, including sapa1Δ (YSBA77), sapa2Δ (YSBA81), sapa3Δ (YSBA119), sapa4Δ (YSBA135), sapa5Δ (YSBA125), sapa6Δ (YSBA91), and sapa7Δ (YSBA127), were washed with dH2O, resuspended in 1 mL of dH2O, and spotted onto solid YCB-BSA media using 3 µL of the cell suspension. The plates were incubated for 3 days at (A) 30°C, (B) 37°C, and (C) 42°C. The values below each figure represent the average halo thickness, indicative of SAP activity. Three biologically independent experiments were performed and one representative data are shown here. (D) SAP activities of SAPA3 deletion mutant and complemented strains. Three biologically independent experiments were performed and one representative data are shown here. (A to D) Error bars indicate standard deviation. Statistical analysis was performed using one-way ANOVA with Bonferroni’s multiple-comparison test (****P < 0.0001; NS, not significant).