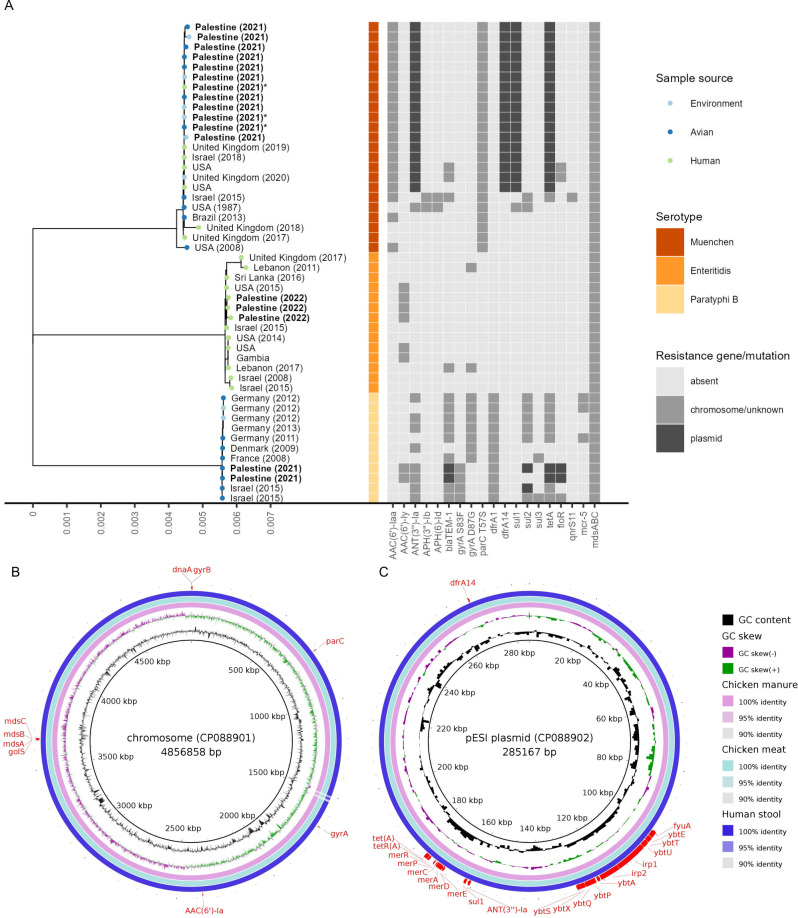
Fig 3.
Phylogenetic analysis of S. enterica isolates. (A) Phylogenetic tree for S. enterica with 17 genomes from Palestine and 31 closely related genomes belonging to the three serotypes found in this study from different countries collected between 1987 and 2020. The presence of AMR markers is shown in light gray if located in the genome or an unidentified plasmid or dark gray if located on a plasmid. Sample sources are depicted by colored, filled circles. Genomes generated in this study are in bold text. Genomes used for Fig. 3B and C are marked with an asterisk. Comparison of Palestinian S. enterica serotype Muenchen chromosome (B) and pESI plasmid (C) from chicken manure and human feces as well as meat with a clinical isolate from Israel collected in 2018. The presence and location of antibiotic-resistance genes are shown in red. The human isolate collected in 2021 is from a broiler chicken production chain worker, while all three human isolates collected in 2022 are from hospitalized gastroenteritis patients.