Figure 2: Fusing de novo designed JDMs with substrate binding domains dissolves intracellular condensates.
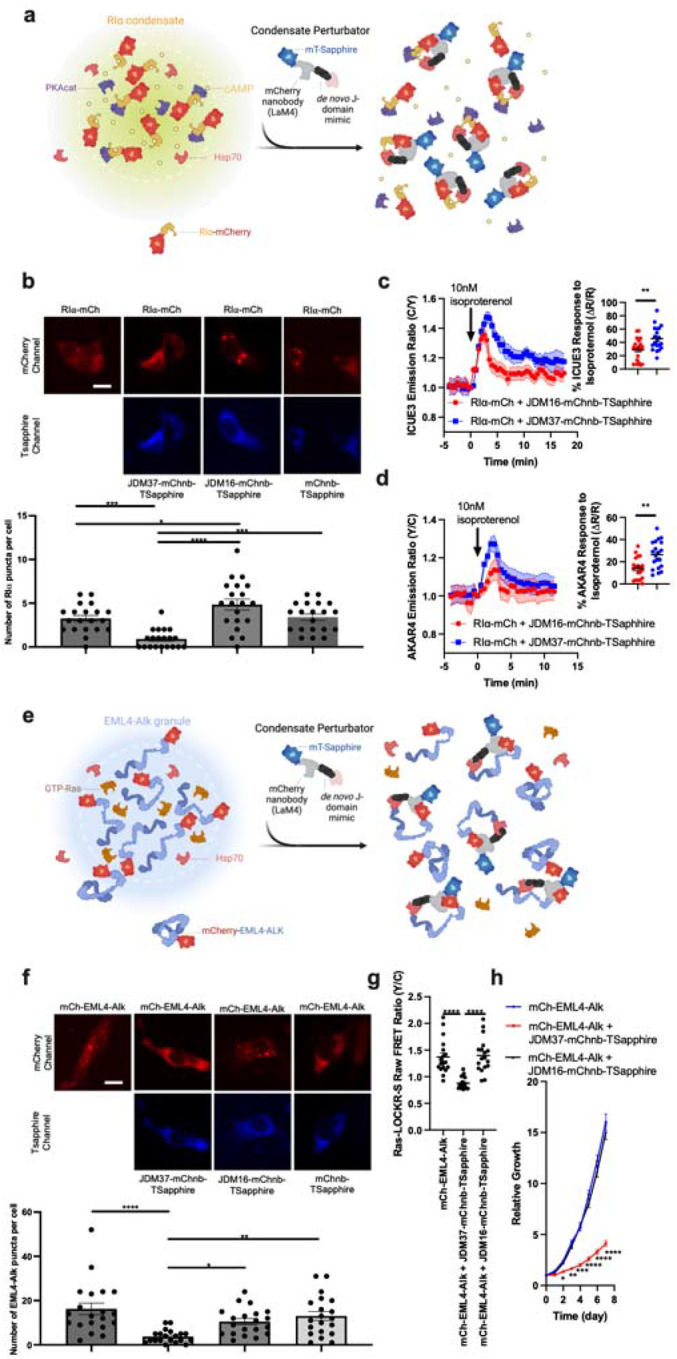
a, Schematic of strategy to dissolve RIα condensates. Protein that drives condensation (here, RIα) is tethered with mCherry. JDM37 is fused to an mCherry nanobody and mT-Sapphire, and this tool is called condensate perturbator. b, Expression of engineered condensate perturbators dissolves mCherry-tagged RIα puncta in HEK293T cells (see Methods for details). Top: Representative epifluorescence images of the various conditions tested. Scale bar = 10μm. Bottom: Quantification of number of RIα puncta per cell. Each point represents a single cell (n=20 cells). c-d, Time-course imaging of HEK293T cells expressing mCherry-tagged RIα, either cAMP sensor ICUE3 (c) or PKA sensor AKAR4 (d), and either JDM37-based or JDM16-based condensate perturbators. In each condition, 10nM isoproterenol was added. Solid lines indicate representative average time with error bars representing standard error mean (SEM) (n=at least 15 cells per curve). e, Schematic of strategy to dissolve EML4-Alk oncogenic condensates. Protein that drives condensation (here, EML4-Alk) is tethered with mCherry. f, Expression of engineered condensate perturbators dissolves mCherry-tagged EML4-Alk puncta in Beas2B cells (see Methods for details). Top: Representative epifluorescence images of the various conditions tested. Scale bar = 10μm. Bottom: Quantification of number of EML4-Alk puncta per cell. Each point represents a single cell (n=20 cells). g, Raw FRET ratios of HEK293T cells expressing mCherry-tagged RIα, Ras sensor Ras-LOCKR-S, and either JDM37-based or JDM16-based condensate perturbators. Each point represents a single cell (n=18 cells). h, Cell growth curves of Beas2B cells expressing mCherry-tagged EML4-Alk with or without condensate perturbators (n=3 experiments). Line represents average from all 3 experiments. For the quantification of number of puncta per cell in b and f, cells only with sufficient expression of the condensate perturbator were chosen for analysis (see Methods for details).
