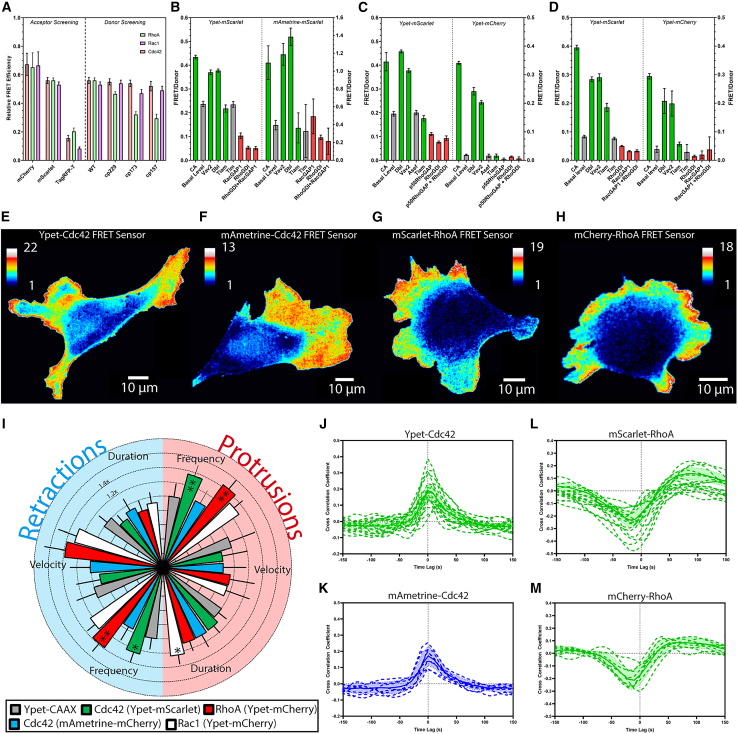
Figure 2.
Development and testing of red-shifted GTPase biosensors. (A) Donor and acceptor optimization for dual-chain FRET GTPase biosensors (n ≥ 3). FRET efficiency of the Cdc42 (B), RhoA (C), and Rac1 (D) biosensors alone (Basal Level, gray) or when exposed to upstream activators (green), inhibitors (red), upstream regulators not specific for the tested GTPase (gray), and activating point mutations (green, CA) (error bars SD, n ≥ 3). Activity maps of Cdc42 (E and F) and RhoA (G and H) biosensors in randomly moving MEFs. Pseudocolor scales indicate ratio values relative to the lowest values (materials and methods). (I) Radial plot summarizing parameters of protrusion (light red, right side) and retraction (light blue, left side) from MEFs expressing biosensors, normalized to Ypet-CAAX (negative control) (error bars 95% CI). Data statistically different from the negative control are marked as ∗ (p-value < 0.05) and ∗∗ (p-value < 0.01); see additional information and alternate representation in Fig. S1 in the supporting material. (J–M) Cross correlation of biosensor activity and edge velocity, showing the lag between changes in protrusion velocity and GTPase activation, for Cdc42 (J and K) and RhoA (L and M) (n ≥ 7, error bands 95% CI).