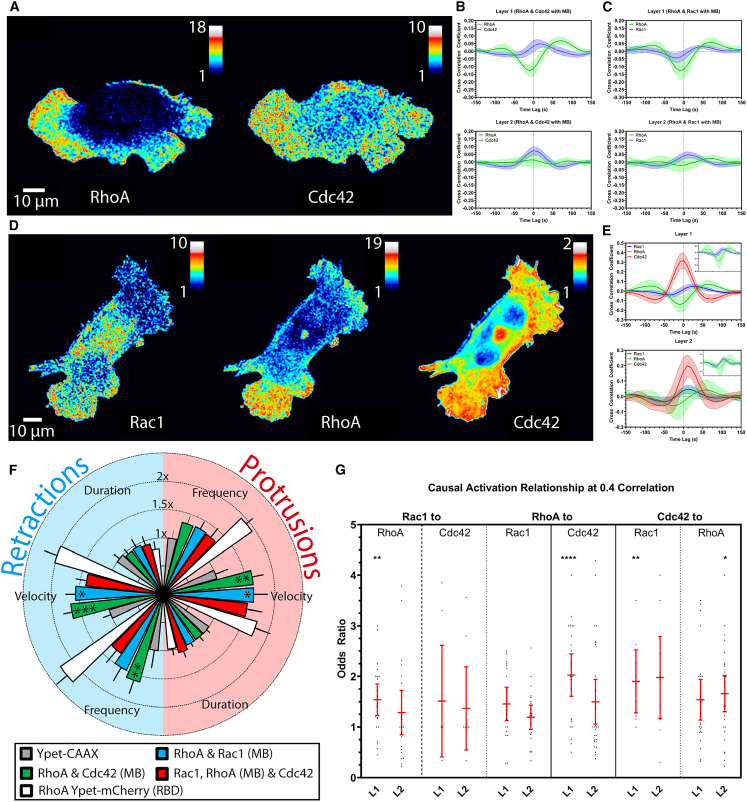
Figure 4.
Application of MultiBinder to imaging and analyzing multiplex signals in the same cell. (A and B) RhoA and Cdc42 activity maps and cross correlations plots for randomly migrating MEFs, obtained using Ypet-RhoA, mAmetrine-Cdc42, and MB-Cherry (n = 17). (C) Cross correlation plots for simultaneously imaged RhoA and Rac1 activities (n = 13). (D and E) Rac1, RhoA, and Cdc42 activity mapped in the same MEF using Ypet-RhoA, mAmetrine-Rac1, MB-Cherry, and the SNAPsense biosensor (n = 8). Cross correlation analyses in (B), (C), and (E) were performed in two layers of probing windows of 1.3 μm width. Layer 1 covered 0–1.3 μm from the cell edge, and Layer 2 covered 1.3–2.6 μm from the edge. (F) Radial plot of protrusion (light red, right side) and retraction (light blue, left side) parameters normalized to the negative control Ypet-CAAX (n = 11) for dual and triple imaging with Ypet-RhoA mCherry-RBD as a positive control (statistical comparisons shown to negative control; see Fig. S8 for full analysis). Error bands 95% CI for (B), (C), and (E). (G) Odds ratio values of GTPase activation events at 0.4 correlation threshold from combined dual and triple imaging datasets (error bars SD).