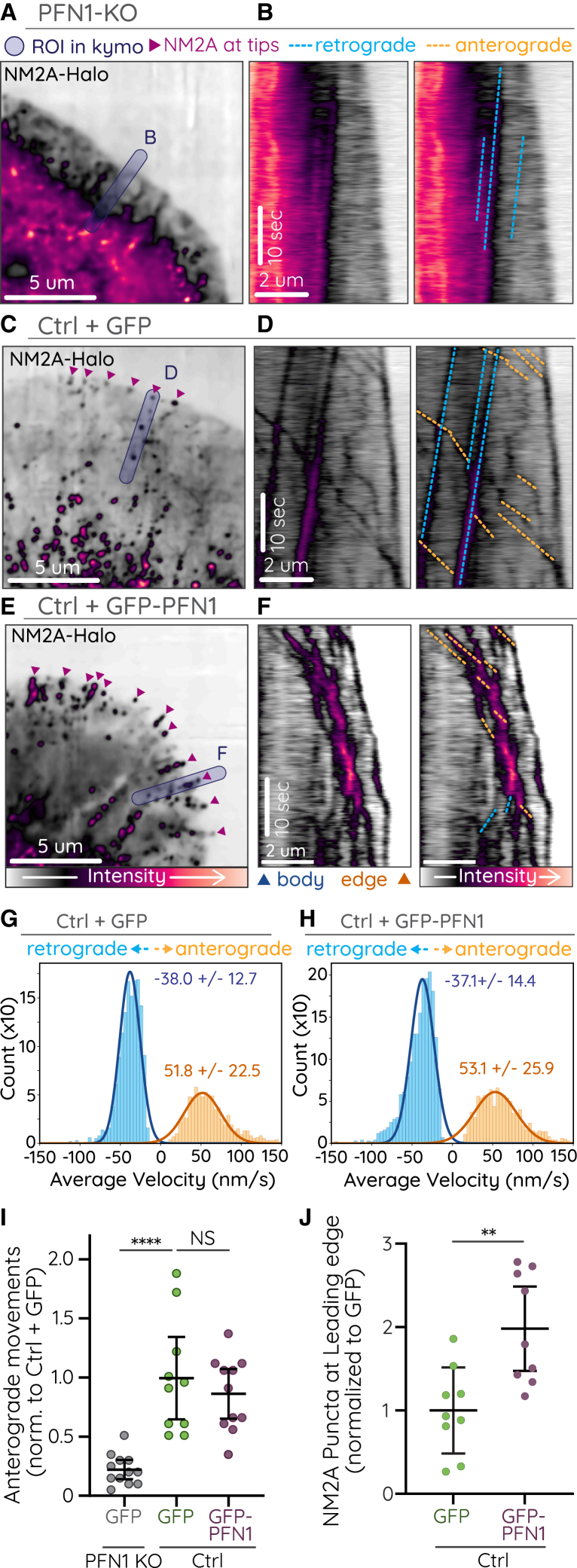
Figure 4.
Processive NM2 is dependent on lamellipodia architecture
NM2A-Halo was imaged in PFN1-KO CAD cells, CAD cells expressing GFP, or CAD cells overexpressing GFP-PFN1. (A–F) Representative images (A, C, E) and duplicate kymographs (B, D, F) with (right) and without (left) annotation of anterograde (orange dotted lines) and retrograde (blue dotted lines) NM2A movements in PFN1 KO (A and B), ctrl + GFP (C and D) and ctrl + GFP-PFN1 (E and F) cells. (G and H) Histograms of velocity measurements from automated tracking of NM2A-Halo puncta in control and PFN1 overexpressing cells (see Table 1). (I) Plot of the number of NM2A-Halo anterograde movements in the lamellipodia protrusions of PFN1 KO, control, and PFN1 overexpressing cells. Only anterograde events that initiated within 8 m of the leading edge were counted. Each dot indicates one cell. Error bars depict mean ± SD. p-values were calculated from a one-way ANOVA followed by Tukey's multiple comparisons post-hoc test. ∗∗∗∗, p <0.0001; NS, not significant. (J) Number of NM2A-Halo puncta within 1 m of the leading edge of PFN1 KO, control, and PFN1 overexpressing cells. Each dot indicates one cell. Error bars depict mean ± SD. p-values were calculated from a student's t-test. ∗∗, p < 0.01. Imaging was performed on Nikon CSU-W1 SoRa spinning disk.