Abstract
The crown bridge loop is hexapeptide motif in which the backbone carbonyl group at position 1 is hydrogen bonded to the backbone imino groups of positions 4 and 6. Residues at positions 1 and 4–6 are held in a tight substructure, but different orientations of the plane of the peptide bond between positions 2 and 3 result in two conformers: the 2,3-αRαR crown bridge loop — found in approximately 7% of proteins — and the 2,3-βRαL crown bridge loop, found in approximately 1–2% of proteins. We constructed a relational database in which we identified 60 instances of the 2,3-βRαL conformer, and find that about half occur in enzymes of the haloacid dehalogenase (HAD) superfamily, where they are located next to the catalytic aspartate residue. Analysis of additional enzymes of the HAD superfamily in the extensive SCOPe dataset showed this crown bridge loop to be a conserved feature. Examination of available structures showed that the 2,3-βRαL conformation — but not the 2,3-αRαR conformation — allows the backbone carbonyl group at position 2 to interact with the essential Mg2+ ion. The possibility of interconversion between the 2,3-βRαL and 2,3-αRαR conformations during catalysis is discussed.
Graphical abstract
Highlights
-
•
The active site of haloacid dehalogenases is supported by a conserved highly hydrogen-bonded 2,3-βRαL crown bridge loop.
-
•
The 2,3-βRαL crown bridge loop occurs infrequently in proteins, but half are found in haloacid dehydrogenases.
-
•
It is possible that the 2,3-βRαL and the 2,3-αRαR conformations of the motif might interconvert during catalysis.
Abbreviations
- HAD
haloacid dehalogenase
The ranges of dihedral angles represented by the different abbreviations are as follows
- αR
–140° < φ < –20°, –90° < ψ < 40°
- sub-αR
–170° < φ < –80°, –160° < ψ < –10°
- αL
20° < φ < 140°, –40° < ψ < 90°
- βR
150° < φ or φ < –25°, 40° < ψ or ψ < –150°
- β L
20° < φ < 140°, –180° < ψ < –80°
1. Introduction
Hydrogen bonds between backbone CO and NH groups are important determinants of the conformation of proteins. This is true not only for secondary structure elements, such as the α-helix and β-sheet with their repeating units, but also for single small structural motifs, such as the β-turn (Venkatachalam, 1968; Richardson, 1981). For some years we have been interested in these small motifs, and our work has been directed at finding what roles they play in proteins (Milner-White and Poet, 1987; Wan and Milner-White, 1999; Torrance et al., 2009; Leader and Milner-White, 2021a).
One class of motifs is characterized by a tripeptide in which the N1 and N3 imino groups form hydrogen bonds with a single anionic group. The anionic group bound by such motifs can be part of a non-protein cofactor or prosthetic group (e.g. the S of iron–sulphur clusters) or a δ–ve oxygen from another part of the polypeptide chain. Only two types of motif have been found that include such N1 and N3 imino group bridges (Leader and Milner-White, 2015). The first, the ‘nest’ (Watson and Milner-White, 2002; Hayward et al., 2014), has attracted interest in relation to a possible role in metalloenzymes during early evolution (Milner-White and Russell, 2008; Nitschke et al., 2013). The second, the ‘crown’, was described more recently (Leader and Milner-White, 2015) and has been less studied. It is the subject of this report.
The crown itself (Fig. 1A) is a tripeptide with the conformation at positions 1 and 2 being what we term sub-αR and αR, respectively. (The term sub-αR was coined (Leader and Milner-White, 2015) to describe a more diffuse region of the Ramachandran plot at lower values of φ and ψ than αR — see Abbreviations.) The tripeptide lies upon an imaginary ring which, if completed, would contain four amino acid residues. The peptide imino groups in positions 1 and 3 have the potential to bridge the same negatively-charged group, and motifs that have been found to be bridged by such a negatively-charged atom (Fig. 1B) are termed crown bridges. (Although we identify fewer examples of crown bridges in our database than of the unbridged crown, it is possible that the δ–ve oxygen of a water molecule acts as the bridging anion in the absence of other groups.) A subset of crown bridges is found in which the anionic group is the backbone carbonyl group located three residues N-terminal to the tripeptide, and the six-residue motif encompassing these positions has been designated the crown bridge loop (Fig. 1C). Only residues 1, 4, and 6 in the crown bridge loop are necessarily involved in the bridging interaction, allowing two different conformations for residues 2 and 3. These conformations define two subcategories, which we refer to as 2,3-αRαR and 2,3-βRαL. (Residues 1–4 actually constitute a β-turn, and 2,3-αRαR and 2,3-βRαL correspond to what are known as ‘type I’ and ‘type II’ conformations (Wilmot and Thornton, 1990).)
Fig. 1.
Two-dimensional diagrammatic representation of different crown structures. (A) The crown. The arrows indicate the orientation of the peptide-bond NH groups. (B) The crown bridge. The square represents a negatively charged ligand bridging the two NH groups. (C) The crown bridge loop. The alternative conformations, 2,3-αRαR (blue lettering) and 2,3-βRαL (red lettering), are indicated. In all frames the numbered circles represent residues centred on the α-carbon atom, with the small red and blue filled circles representing peptide-bond CO and NH groups, respectively. Numbering is from the N-terminus of the peptide, so that corresponding residues are assigned different numbers in the hexapeptide of C from those in the tripeptide of A and B. They have been coloured and presented in the same manner to make their relationship clear. Dihedral angles, αR etc., are defined in Abbreviations. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The crown motif occurs relatively infrequently, and our original database of approximately 400 proteins (https://motif.mvls.gla.ac.uk/motif/index.html) contains only 27 examples of crown bridge loops with the 2,3-αRαR conformation, and three with the 2,3-βRαL conformation. To address the role of these motifs, a larger dataset was needed. This work describes the analysis of motifs in such a dataset, and demonstrates a role for the crown bridge loop in the haloacid dehalogenase family of proteins.
2. Methods
The general approach used to prepare a dataset of a particular motif was: (1) to construct a new, larger, relational database of the structural features of proteins, (2) query this with descriptions of hydrogen-bonded small motifs, and add the retrieved motifs to the database as secondary data, (3) add SCOPe architectural classifications (Chandonia et al., 2017) to the database, and (4) for a particular motif of interest containing a specific SCOPe fold, construct an expanded dataset using additional examples from the SCOPe web resource. The details are as follows.
-
(1)
The Richardson laboratory ‘Top 8000’ was the source of the three-dimensional protein structures used in this work (http://kinemage.biochem.duke.edu/research/top8000/). It consists of high-resolution, quality-filtered, individual protein subunits which are 70% non-redundant in terms of structure. These structures were processed programmatically (Leader and Milner-White, 2009) allowing a MySQL relational database to be populated with the essential information required to describe motifs: coordinates of atoms, identities and dihedral angles of residues, descriptors of proteins, and characteristics of hydrogen bonds. The final database (ProtMotif2) contains information from 4484 individual protein subunits.
-
(2)
Queries employing the Structured Query Language (SQL) were used to retrieve motifs of various types, which were then included in ProtMotif2 as secondary data. (For example, for the 2,3-βRαL crown bridge loop — Fig. 1C — the query specifies six contiguous residues in which there are hydrogen bonds from the backbone CO of residue 1 to the backbone NHs of residues 4 and 6, and conformations βR and αL at positions 2 and 3, respectively.).
-
(3)
In previous work on a different motif, the β-link (Leader and Milner-White, 2021b), the SCOPe architectural classification of protein structures was found useful for identifying the types of protein containing the motif and its location therein. We therefore incorporated SCOPe data (from file dir.cla.scope.2.07-stable.txt at https://scop.berkeley.edu) into database tables, allowing individual motifs to be related to the SCOPe fold in which they occur. (Some proteins in the ProtMotif2 database are not represented in the SCOPe data file, and could not be assigned a SCOPe classification.) A publicly available web application, Protein Motif 2 (https://motif.mvls.gla.ac.uk/ProtMotif21/), was constructed to allow access to this information (Fig. S1).
-
(4)
We used these resources to identify SCOPe folds in which the motif of interest occurred — fold c.108, in the case of the 2,3-βRαL crown bridge loop. We then expanded the dataset for this fold (from 21 to the 60 in Table S2(i)) by selecting more instances from the SCOPe web resource. As fold c.108 contains twenty five families, c.108.1.1 to c.108.1.25, the primary consideration was to ensure all these families were represented. Other criteria for selection were: resolution, inclusion of ligand, characterization and species origin. Selected structures were examined in Jmol (Herráez, 2006) for hydrogen-bonding patterns, and analysed using the program, DSSP (Kabsch and Sander, 1983) to determine dihedral angles.
3. Results
The newly constructed ProtMotif2 database of structural information for 4484 protein subunits was queried for crown bridge loops, and 313 were retrieved: 253 of type 2,3-αRαR and 60 of type 2,3-βRαL. The increased representation of the latter type (previously only three examples had been identified) allowed a broad analysis with respect to SCOPe architectural classification as described in Methods. Crown bridge loops of type 2,3-αRαR in the database were found in many different folds, with none containing more than six individual examples. In contrast, a single fold — SCOPe class c.108 — was assigned to more than half of the 2,3-βRαL crown bridge loops (21 of 38 instances). This fold contains a six-stranded β-sheet, and in each case the 2,3-βRαL crown bridge loop is found at the C-terminal end of the third strand of the sheet, allowing it to be located easily despite a variation in the overall structure of proteins of this family (Fig. S2).
Fold c.108 is described by SCOPe as ‘HAD-like’, meaning that it contains members of the haloacid dehalogenase (HAD) superfamily. Using the SCOPe web resource (https://scop.berkeley.edu), we examined other members of the fold, including sub-families not represented in the ProtMotif2 database, to determine whether this crown bridge loop is a feature of other HAD-family proteins. On the basis of measurement of dihedral angles and calculation of hydrogen bonds we determined that 60 of 63 examples contain a 2,3-βRαL crown bridge loop. The three apparent exceptions are all predicted by the AlphaFold program to contain a 2,3-βRαL crown bridge loop (see Discussion), and so we consider this motif to be a general feature of HAD-family proteins.
The HAD superfamily was named for its founder members, a class of haloacid dehalogenase (Hisano et al., 1996). However it is actually a large and diverse family of proteins, overwhelmingly composed of phosphohydrolases (phosphatases), and is found in all domains of life (Allen and Dunaway-Mariano, 2009). The phosphatases include ATPases, sugar phosphatases, sugar phosphomutases, phosphoaminoacid phosphatases and deoxyribonucleotidases. In most cases HAD-family enzymes have two structural domains. A core catalytic domain contains a conserved aspartate residue and an essential Mg2+ cofactor. A second domain (absent or rudimentary in some cases) is termed the ‘capping domain’ (see Fig. S2(ii)) and, where present, is involved in substrate recognition (Allen and Dunaway-Mariano, 2009). Different categories of capping subdomain (C0, C1, C2a or C2b) connect to the core catalytic domain at different positions in the polypeptide chain. During catalysis the capping domain moves with respect to the core catalytic domain between a ‘closed’ structure, in which solvent is excluded from the active site, and an ‘open’ structure which allows binding and release of substrate.
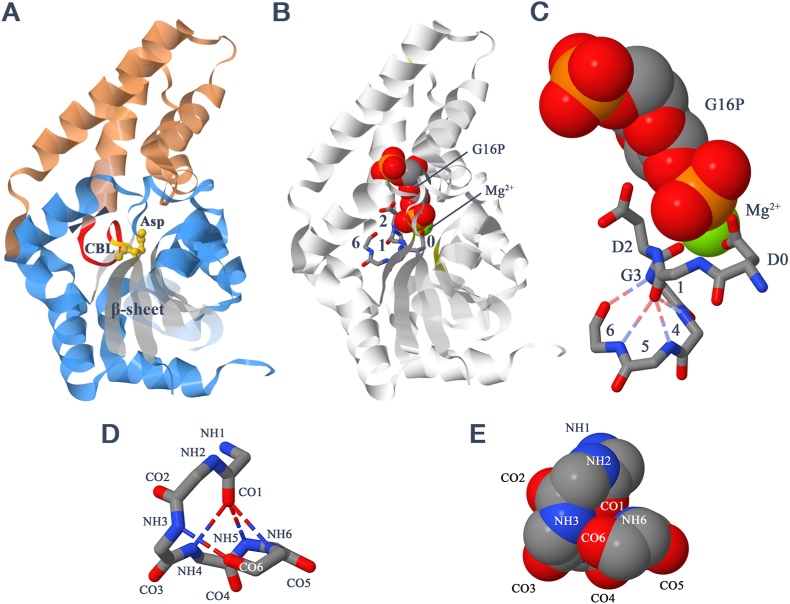
Where is the crown bridge loop located in relation to the catalytic centre of the HAD-family enzymes? Fig. 2A shows its general position in the overall structure of a phosphoglucomutase and its proximity to the catalytic aspartate at the face of the core domain where the latter interacts with the cap domain (Lahiri et al., 2003). In members of the HAD-family with a C1 cap, such as this, the crown bridge loop continues as part of a ‘hinge’ region connecting to an α-helix at the N-terminus of the cap domain. This hinge has been described as consisting of a ‘squiggle’ and a ‘flap’, conserved through the superfamily. The ‘squiggle’ is actually equivalent to the first five residues of the crown bridge loop, and the ‘flap’ is the continuation into the cap domain (Burroughs et al., 2006). A similar view to Fig. 2A, but with the crown bridge loop shown in more detail and the substrate and Mg2+ ion included, is presented in Fig. 2B. Following convention, the catalytic aspartate is referred to as Asp0, and residues C-terminal to it are numbered sequentially preceded by a ‘+’. Asp0 is adjacent to a pentapeptide consensus sequence, in which the most frequent residues (in the structures considered here) are (L/I/M/V)DG(T/V)(L/I) — see Table S1. Positions +1 to +5 of the consensus sequence constitute the first five residues of the crown bridge loop, with the consensus DG (+2 and +3 of the pentapeptide) equivalent to the residues defining the 2,3-βRαL conformation of the latter. Fig. 2C shows an enlarged view of this region of the catalytic centre. It can be seen how the side-chain carboxylate of Asp0 and the backbone carbonyl of Asp+2 chelate the Mg2+ ion. The side-chain carboxylate of Asp+2 is oriented away from the Mg2+ ion, and interacts with a phosphate group of the substrate. (A transition-state analogue is shown here.)
Fig. 2.
Active site of HAD-family enzymes. (A) General view of β-phosphoglucomutase from Lactococcus lactis (PDB entry 1o03 (Lahiri et al., 2003)) as a ribbon diagram, but with the catalytic aspartate as ball-and-stick. The cap domain is coloured salmon pink and the catalytic core domain light blue, except for the six-stranded β-sheet (white), the crown bridge loop (red) and the catalytic aspartate (gold). The start of the linking ‘flap’ is indicated by arrowheads. Residues ‘in front’ of the β -sheet have been rendered translucent. CBL = crown bridge loop. (B) As A, but with the reaction intermediate, glucose 1,6-bisphosphate (G16P), and the Mg2+ ion shown in space-filling mode, and the catalytic aspartate (0) and adjacent crown bridge loop (numbered without the ‘+’) shown as protein backbone. Sidechains are included for aspartates at positions 0 and +2. (C) Enlarged view of portion of B with substrate, Mg2+ ion, catalytic aspartate and adjacent crown bridge loop. The three most highly conserved amino acids are designated D0, D2 and G3. (D0 in PDB entry 1o03 is Asp8.) (D) Hydrogen bonding of the 2,3-βRαL crown bridge loop (residues 10–15) at the catalytic site of the phosphatase domain of the human lipid phosphatase/epoxide hydrolase (PDB entry 1s8o (Gomez et al., 2004)). Only the backbone is shown. CO and NH groups are numbered 1–6 with reference to the catalytic Asp9 as 0. (E) As D, but in space-filling representation. Only the visible residues are labelled. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Residues 3–6 of the 2,3-βRαL crown bridge loop lie ‘below’ Asp0 of the active site, as illustrated in Fig. 2D for the phosphatase domain of the bifunctional lipid phosphatase/epoxide hydrolase from Homo sapiens (Gomez et al., 2004). The backbone imino groups of residues 4–6 form a puckered ring, slightly above which lies the imino group of residue 3. In many cases this ring is completed, as here, by a CO6←NH3 hydrogen bond. One can consider the major part of the crown bridge loop as a slightly distorted pyramid with CO1 as the apex and residues +3 to +6 forming its square base. Its compact nature is illustrated by the space-filling representation in Fig. 2E. Additional hydrogen bonds would appear to stabilize the substructure. The extra bonds between atoms CO1←NH5 and CO6←NH3 in this example are quite common (Table S2). Less frequently there is an additional hydrogen bond between atoms CO0←NH5.
As mentioned above, the conserved 2,3-βRαL crown bridge loop in HAD-family enzymes is equivalent to the previously described ‘squiggle’. It has been suggested (Burroughs et al., 2006) that the ‘squiggle’ might be an example of a single turn of the rare π-helix (Weaver, 2000; Cooley et al., 2010), which is defined by repeating hydrogen bonds between residues i ← i + 5 (1←6 in the crown bridge loop — Fig. 1C). An essential feature of an oligopeptide helix is the repetition of dihedral angles, producing a similar non-zero rise, d, between residues (Dickerson and Geis, 1969). Here, in contrast, positions +4 to +6 lie on a flat ring (Fig. 2D). Moreover the dihedral angles in the 2,3-βRαL crown bridge loop vary markedly — successively αR, βR, αL, sub-αR, αR, βR — clearly differentiating it from the π-helix, in which dihedral angles should lie in the bottom left-hand quadrant of the Ramachandran plot (see Fig. S3).
A pertinent question is whether the crown-bridge loop or ‘squiggle’ changes conformation as the cap and core domains move relative to one another during the transition from the ‘cap-closed’ to the ‘cap-open’ structure. Dai et al. noted “a change in the backbone φ and ψ angles of hinge residues Thr14, Asp15, Thr16 and Ala17” of β-phosphoglucomutase, equivalent to positions +6 to +9 in the ‘Asp0’ convention (Dai et al., 2009). This has been interpreted by another group as “unwinding of the squiggle element” (Park et al., 2015). We therefore examined the structural data for the 2,3-βRαL crown bridge loop in β-phosphoglucomutase trapped in the cap-open form (Lahiri et al., 2002), rather than the cap-closed form that was shown in Fig. 2. It can be seen from Fig. 3 that although the overall structure of the protein has changed, and there has indeed been an alteration in the backbone ψ angle at position +6 (Thr14), the hydrogen-bonds of the crown bridge loop — including the 1←6 bond — have been retained. Similar retention of the hydrogen bonding of the crown bridge loop is also seen in the sucrose-phosphate phosphatase from the cyanobacterium Synechocystis sp PCC 6803 (Fieulaine et al., 2005) (Fig. S4).
Fig. 3.
Comparison of conformation of the crown bridge loop in the cap-open and cap-closed forms of phosphoglucomutase. Backbone residues 8–14 and hydrogen bonds in β-phosphoglucomutase from Lactococcus lactis. (A) Cap-closed form (PDB entry 1o03 (Lahiri et al., 2003)). (B) Cap-open form (PDB entry 1lvh (Lahiri et al., 2002)). (C) Superimposition of closed (red and orange) and open (blue and light blue) forms. (D) Dihedral angles of the crown bridge loop (residues 9–14). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
The crown bridge loops at the active site of HAD-family enzymes are predominantly in the 2,3-βRαL, rather than the 2,3-αRαR, conformation. This is consistent with the high conservation at position +3 of the amino acid, glycine, which is able to occupy the generally unfavourable αL region of the Ramachandran plot (see Tables S1 and S3). In the few cases, already mentioned, of structures in which the 2,3-αRαR or another conformation was found, the prediction by AlphaFold (https://alphafold.ebi.ac.uk) is of the 2,3-βRαL conformation (Table S4). Thus, where different structures were observed in different subunits, the 2,3-βRαL conformation may be the more thermodynamically stable ‘default’. In other cases, the observed structure may perhaps be an alternative to this default, favoured by the conditions of crystallization.
Why has the 2,3-βRαL crown bridge loop been so highly conserved? Residues 4–6 of the loop form a tight substructure with residue 1, which is flanked by catalytic Asp0 and Asp+2 (D0 and D2 in Fig. 2B). In general this would appear to facilitate the interaction of the first aspartate side chain with the Mg2+ ion and the second with the substrate phosphate group. Specifically, the structure allows a functional interaction between the backbone carbonyl group at position 2 (CO2) and the essential Mg2+ ion, which would, in turn, be likely to stabilize the interaction of the ion with the sidechain of Asp0. Further stabilization will occur where there is a hydrogen bond of the type mentioned above between the backbone carbonyl of Asp0 (CO0) and the backbone imino group NH5 (Table S2(i)).
The 2,3-βRαL conformation of the crown bridge loop can be seen as a requirement for the interaction of CO2 and the Mg2+ ion, just described. Let us considered this in more detail. Fig. 4A shows a diagram of the first four residues of (i) a 2,3-βRαL and (ii) a 2,3-αRαR crown bridge loop, without the side-chains but showing the 1←4 hydrogen bond. The difference between the two structures is essentially the rotation of the plane of the peptide bond linking positions +2 and +3 (arrowhead) through approximately 160°, which causes a major movement of the backbone carbonyl group of Asp+2 (CO2 in the figure). The co-ordinates for this example are taken from the phosphatase domain of the bifunctional murine lipid phosphatase/epoxide hydrolase (Argiriadi et al., 1999). In this homodimeric structure the crown bridge loop of one subunit (B in PDB entry 1cr6) has the 2,3-βRαL conformation, whereas the second subunit (A) has the 2,3-αRαR conformation. Fig. 4B shows the complete crown bridge loop and catalytic aspartate for these two conformers. There is a clear difference between them in the position of CO2 and that of the Asp+2 side-chain, which in the 2,3-αRαR conformer is no longer in the proximity of Asp0 and hence is no longer directed towards the substrate. It should be noted that, as far as can be judged, the overall structures of these two subunits are similar (Fig. S5). A second example of this type, a phosphoglycolate phosphatase from Thermoplasma acidophilum, is shown in Fig. 4C (Kim et al., 2004). This illustrates the interaction of Asp0 and CO2 with the metal ion (Ca2+ taking the place of Mg2+) in the 2,3-βRαL conformer, although the overall situation is complicated by the fact that there are two metal ions near the active site in the 2,3-αRαR conformer. The change in orientation at Asp+2 is also apparent.
Fig. 4.
The crown bridge loop of HAD family enzymes in the 2,3-βRαL and 2,3-αRαR conformations. (A) Diagram of backbone of residues +1 to +4 of a crown bridge loop showing the peptide planes as yellow rectangles. (i) 2,3-βRαL conformation, (ii) 2,3-αRαR conformation. The arrowhead indicates the peptide plane between Asp+2 and Gly+3. The co-ordinates are taken from the phosphatase domain of the murine lipid phosphatase/epoxide hydrolase, PDB entry 1cr6 (Argiriadi et al., 1999)). (B) As A, but representation of the complete heptapeptide (residues 9–15), labelled as in Fig. 2, with the side-chains of Asp0 and Asp+2 (only) shown. (i) 1cr6_B — 2,3-βRαL conformation, (ii) 1cr6_A — 2,3-αRαR conformation. (C) Phosphoglycolate phosphatase from Thermoplasma acidophilum, PDB entry 1l6r (Kim et al., 2004), residues 8–14. The labels are as in B, with the Ca2+ ions (spacefill) green. (i) 1l6r_B — 2,3-βRαL conformation, (ii) 1l6r_A — 2,3-αRαR conformation. The diagram have been prepared so that the backbone at positions 0–1 and 4–5 aligns in the two parts. The distances between the closest oxygen atom of Asp0 and that of CO2 in (i) and (ii) are 2.93 Å and 5.21 Å, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Although the 2,3-βRαL conformation of the crown bridge loop in HAD-family enzymes would seem to be that required for catalysis, the two cases just mentioned indicate that adoption of the 2,3-αRαR conformation is energetically feasible, and suggest that it could occur in certain circumstances in vivo. This raises the intriguing possibility that the crown bridge loop might alternate between the 2,3-βRαL and 2,3-αRαR conformations during catalysis, for example to allow release of products and movement of the metal cofactor (Allen and Dunaway-Mariano, 2004). This type of change in conformation is known as ‘peptide-plane flipping’, and has previously been described for β-turns of the type that the four residues in Fig. 4A constitute (Gunasekaran et al., 1998; Hayward, 2001). There is at least one precedent for peptide-plane flipping in an enzyme reaction mechanism: that of residues 58–59 of flavodoxin when it is reduced to its semiquinone form (Alagaratnam et al., 2005). It should be emphasized that there is, as yet, no direct evidence for peptide-plane flipping in HAD-family enzymes, and that in both the cap-open and cap-closed forms the crown bridge loop is found in the 2,3-βRαL conformation (Fig. 3D). Nevertheless, such a dynamic role for the 2,3-βRαL crown bridge loop — in addition to its static one in orienting the Asp+2 carbonyl group — remains an intriguing possibility, and might help to explain the extreme conservation of this ancient structure in HAD-family enzymes.
CRediT authorship contribution statement
David P. Leader: Conceptualization, Methodology, Software, Investigation, Writing – original draft, Writing – review & editing, Visualization. E. James Milner-White: Conceptualization, Methodology, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Handling Editor: Dr. N Strynadka
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2023.100105.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
The data were obtained from our public Motivated Proteins 2 web application which is freely accessible without restriction.
References
- Alagaratnam S., van Pouderoyen G., Pijning T., Dijkstra B.W., Cavazzini D., Rossi G.L., Van Dongen W.M.A.M., van Mierlo C.P.M., van Berkel W.J.H., Canters G.W. A crystallographic study of Cys69Ala flavodoxin II from Azotobacter vinelandii: structural determinants of redox potential. Protein Sci. 2005;14:2284–2295. doi: 10.1110/ps.051582605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen K.N., Dunaway-Mariano D. Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem. Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Allen K.N., Dunaway-Mariano D. Markers of fitness in a successful enzyme superfamily. Curr. Opin. Struct. Biol. 2009;19:658–665. doi: 10.1016/j.sbi.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiriadi M.A., Morisseau C., Hammock B.D., Christianson D.W. Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10637–10642. doi: 10.1073/pnas.96.19.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs A.M., Allen K.N., Dunaway-Mariano D., Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- Chandonia J.-M., Fox N.K., Brenner S.E. SCOPe: manual curation and artifact removal in the structural classification of proteins – extended database. J. Mol. Biol. 2017;429:348–355. doi: 10.1016/j.jmb.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley R.B., Arp D.J., Karplus P.A. Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J. Mol. Biol. 2010;404:232–246. doi: 10.1016/j.jmb.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Finci L., Zhang C., Lahiri S., Zhang G., Peisach E., Allen K.N., Dunaway-Mariano D. Analysis of the structural determinants underlying discrimination between substrate and solvent in β-phosphoglucomutase catalysis. Biochemistry. 2009;48:1984–1995. doi: 10.1021/bi801653r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R.E., Geis I. Harper and Row; New York: 1969. The Structure and Action of Proteins. [Google Scholar]
- Fieulaine S., Lunn J.E., Borel F., Ferrer J.-L. The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell. Plant Cell. 2005;17:2049–2058. doi: 10.1105/tpc.105.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez G.A., Morisseau C., Hammock B.D., Christianson D.W. Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis. Biochemistry. 2004;43:4716–4723. doi: 10.1021/bi036189j. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K., Gomathi L., Ramakrishnan C., Chandrasekhar J., Balaram P. Conformational interconversions in peptide β-turns: analysis of turns in proteins and computational estimates of barriers. J. Mol. Biol. 1998;284:1505–1516. doi: 10.1006/jmbi.1998.2154. [DOI] [PubMed] [Google Scholar]
- Hayward S. Peptide-plane flipping in proteins. Protein Sci. 2001;10:2219–2227. doi: 10.1110/ps.23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S., Leader D.P., Al-Shubailly F., Milner-White E.J. Rings and ribbons in protein structures: characterization using helical parameters and Ramachandran plots for repeating dipeptides. Proteins. 2014;82:230–239. doi: 10.1002/prot.24357. [DOI] [PubMed] [Google Scholar]
- Herráez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- Hisano T., Hata Y., Fujii T., Liu J.Q., Kurihara T., Esaki N., Soda K. Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An α/β hydrolase structure that is different from the α/β hydrolase fold. J. Biol. Chem. 1996;271:20322–20330. doi: 10.1074/jbc.271.34.20322. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kim Y., Yakunin A.F., Kuznetsova E., Xu X., Pennycooke M., Gu J., Cheung F., Proudfoot M., Arrowsmith C.H., Joachimiak A., et al. Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum. J. Biol. Chem. 2004;279:517–526. doi: 10.1074/jbc.M306054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S.D., Zhang G., Dunaway-Mariano D., Allen K.N. Caught in the act: the structure of phosphorylated β-phosphoglucomutase from Lactococcus lactis. Biochemistry. 2002;41:8351–8359. doi: 10.1021/bi0202373. [DOI] [PubMed] [Google Scholar]
- Lahiri S.D., Zhang G., Dunaway-Mariano D., Allen K.N. The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction. Science. 2003;299:2067–2071. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- Leader D.P., Milner-White E.J. Motivated proteins: a web application for studying small three-dimensional protein motifs. BMC Bioinf. 2009;10:60. doi: 10.1186/1471-2105-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader D.P., Milner-White E.J. Bridging of partially negative atoms by hydrogen bonds from main-chain NH groups in proteins: the crown motif. Proteins. 2015;83:2067–2076. doi: 10.1002/prot.24923. [DOI] [PubMed] [Google Scholar]
- Leader D.P., Milner-White E.J. Identification and characterization of two classes of G1 β-bulge. Acta Crystallogr. 2021;D77:217–223. doi: 10.1107/S2059798320015533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader D.P., Milner-White E.J. The β-link motif in protein architecture. Acta Crystallogr. 2021;D77:1040–1049. doi: 10.1107/S2059798321006768. [DOI] [PubMed] [Google Scholar]
- Milner-White E.J., Poet R. Loops, bulges, turns and hairpins in proteins. Trends Biochem. Sci. 1987;12:189–192. [Google Scholar]
- Milner-White E.J., Russell M.J. Predicting the conformations of peptides and proteins in early evolution. Biol. Direct. 2008;3:3. doi: 10.1186/1745-6150-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke W., McGlynn S.E., Milner-White E.J., Russell M.J. On the antiquity of metalloenzymes and their substrates in bioenergetics. Biochim. Biophys. Acta. 2013;1827:871–881. doi: 10.1016/j.bbabio.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Park J., Guggisberg A.M., Odom A.R., Tolia N.H. Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1. Acta Crystallogr. 2015;D71:1824–1834. doi: 10.1107/S1399004715012067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J.S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Torrance G.M., Leader D.P., Gilbert D.R., Milner-White E.J. A novel main chain motif in proteins bridged by cationic groups: the niche. J. Mol. Biol. 2009;385:1076–1086. doi: 10.1016/j.jmb.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Venkatachalam C.M. Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers. 1968;6:1425–1436. doi: 10.1002/bip.1968.360061006. [DOI] [PubMed] [Google Scholar]
- Wan W.-Y., Milner-White E.J. A recurring two-hydrogen-bond motif incorporating a serine or threonine residue is found both at α-helical N termini and in other situations. J. Mol. Biol. 1999;286:1651–1662. doi: 10.1006/jmbi.1999.2551. [DOI] [PubMed] [Google Scholar]
- Watson J.D., Milner-White E.J. A novel main-chain anion-binding site in proteins: the nest. A particular combination of φ,ψ values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 2002;315:171–182. doi: 10.1006/jmbi.2001.5227. [DOI] [PubMed] [Google Scholar]
- Weaver T.M. The π-helix translates structure into function. Protein Sci. 2000;9:201–206. doi: 10.1110/ps.9.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmot C.M., Thornton J.M. β-turns and their distortions: a proposed new nomenclature. Protein Eng. 1990;3:479–493. doi: 10.1093/protein/3.6.479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data were obtained from our public Motivated Proteins 2 web application which is freely accessible without restriction.