Abstract
SATB1 (Special A-T rich Binding protein 1) is a cell type-specific factor that regulates the genetic network in developing T cells and neurons. In T cells, SATB1 is required for lineage commitment, VDJ recombination, development and maturation. Considering that its expression varies during B-cell differentiation, the involvement of SATB1 needs to be clarified in this lineage. Using a KO mouse model in which SATB1 was deleted from the pro-B-cell stage, we examined the consequences of SATB1 deletion in naive and activated B-cell subsets. Our model indicates first, unlike its essential function in T cells, that SATB1 is dispensable for B-cell development and the establishment of a broad IgH repertoire. Second, we show that SATB1 exhibits an ambivalent function in mature B cells, acting sequentially as a positive and negative regulator of Ig gene transcription in naive and activated cells, respectively. Third, our study indicates that the negative regulatory function of SATB1 in B cells extends to the germinal center response, in which this factor limits somatic hypermutation of Ig genes.
Keywords: B cells, Nuclear factor, Ambivalent, Somatic hypermutation
Subject terms: Immunology, Gene regulation in immune cells
Introduction
The transcription factor Special A-T rich binding protein 1 (SATB1) is a factor able to bind to Matrix Attachment Regions (MAR) in the nucleus [1]. This specific MAR-binding protein (MAR-BP) is indispensable for T lymphocyte development [2–4] through its involvement in properly organizing nuclear architecture, especially chromatin folding [5–7]. Murine SATB1, sharing more than 98% identity with its human homolog, can multimerize through a ubiquitin-like domain [8] and interacts with chromatin through its CUT-like and homeodomains, which are involved in DNA binding affinity and specificity, respectively [9, 10]. One historic feature of SATB1 is its ability to bind nuclear matrix proteins [11, 12]. Another essential feature, as a transcription factor, is its capacity to bind DNA for gene regulation. Strikingly, such interactions were found to occur in nucleosome-dense regions, preferentially at AT-rich sequences in the nucleosomal core [13], a feature that strongly supports its candidacy as a pioneer factor. Given its multivalent potency as a nuclear matrix scaffolding factor, a genome organizer involved in chromatin looping and a transcription factor, the exact mechanism linking this MAR-BP to gene regulation remains puzzling. The large body of literature related to SATB1 in T cells suggests that this factor is capable of flexible functions, either binding to euchromatin [5, 6] or nucleosome-dense regions [13], depending on the cell type and development stage assessed. Long-distance interactions between promoters and enhancers can be promoted by SATB1 homotetramerization [8]. A negative regulatory function for SATB1 was attributed to its capacity to recruit chromatin modifiers such as histone deacetylase 1 (HDAC1) [14]. Indeed, SATB1 is subject to posttranslational modification: phosphorylation of this MAR-BP modifies interactions with chromatin corepressor and coactivator complexes, leading to a switch in its transcriptional activity [15, 16]; SATB1 acetylation disrupts its interaction with C-terminal binding protein 1 (CtBP1) [17]. Moreover, regulation of the Satb1 gene is tightly regulated in T cells by interchanging promoter usage, resulting in fine tuning of protein expression [18, 19]. These multiple levels of regulation suggest the need for meticulous control of SATB1 expression.
SATB1 deletion in mice led to major alterations in neuronal, hematopoietic and immune systems [2, 20] and pointed out broad and critical functions for this protein in mouse development. SATB1 is a major player in early hematopoiesis since it promotes hematopoietic stem cell (HSC) self-renewal [21]. Based on its relative expression level in differentiating HSCs [22], SATB1 favors the lymphoid lineage [23]. In erythrocytes and myeloid cells, this protein also modulates epigenetic marks by association with CREB binding protein (CBP) to control β-globin and NADPH oxidase gene expression [24–26]. There is also growing evidence showing that SATB1 is involved in cancer and autoimmune diseases [6, 7, 27, 28], highlighting the necessity to decipher its regulation in immune cells.
Due to thymus alteration in Satb1 KO mice [2], SATB1’s functions were largely studied in T cells. Initially described as a repressor factor [12, 29], SATB1 proved to also be a transcriptional activator [6, 30]. As an example of ambivalence, SATB1 could repress c-myc in resting T cells while stimulating its expression in activated T cells [3]. It is now established that SATB1 is a major regulator of thymocyte development throughout all differentiation stages from progenitors to regulatory subsets [28, 31, 32]. One notable role of SATB1 in T cells is its activator function for Rag gene expression to promote VDJ recombination and shape the T-cell receptor repertoire [33, 34]. In this context, SATB1 has been shown to bridge Rag1 and Rag2 gene promoters and the distant antisilencer element (ASE) of this locus [33].
In contrast to the vast knowledge on SATB1 fine tuning and regulatory functions in T lymphocytes, little is known about the potency of SATB1 in regulating B-cell development beyond HSC fate decisions [22, 23]. Initial studies reported a discrete defect in B-cell numbers in Satb1 KO mice [2]. Taking advantage of their elegant fluorescent reporter model, the Yokota group described fluctuations in Satb1 expression throughout B-cell development with higher expression in naive B-cell subsets. In this study, the authors proposed that SATB1 is involved in BCR-mediated B-cell survival [35]. Many aspects of the literature point to a potential function for SATB1 during B-cell development. Given its regulatory function in Rag gene expression in T cells [33], the potential implication of SATB1 in V(D)J recombination of Ig genes in B cells needs to be examined. Moreover, since SATB1 was discovered by its ability to bind the MAREµ region of the IgH locus [1], we postulated a role for SATB1 in Ig heavy chain expression.
In this study, we examined SATB1 involvement in B-lineage cell development and depicted its critical function in immunoglobulin production using a conditional KO mouse model allowing deletion of this factor in B-lineage cells. Although we found SATB1 to be nonessential for B-cell development, we showed that this factor displays an ambivalent function in late developing B-cell subsets, acting sequentially as a positive and then negative regulator of Ig gene transcription. The negative regulatory function of SATB1 extends to the germinal center reaction in which this factor limits Ig gene somatic hypermutation.
Materials and methods
Mice
Satb1tm1a/wt mice were obtained from the Mouse Clinical Institute (MCI) (IR00004167/P4167). First, crossings with 129S4/SvJae-Gt(ROSA)26Sortm2(FLP*)sor/J mice (The Jackson Laboratory) removed the reporter and marker cassette placed between Frt sites. Second crossings with B6. C(CG)-Cd79atm1(cre)Reth/EhobJ mice (The Jackson Laboratory) deleted Satb1-exon4 due to CRE recombinase expression in B-lineage cells [36]. Aicda−/− homozygous mice (kindly provided by Pr. T. Honjo) were used to prepare control samples devoid of SHM, as reported in [37]. All experiments, except 3C-HTGTS, were performed on two-month-old mice. The primers used for genotyping are listed in Supplementary Table S1. All animal strains were bred and maintained in SPF conditions at 21–23 °C with a 12-h light/dark cycle. Procedures were reviewed and approved by the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation APAFIS#16639-2018090612249522v2.
Western blotting
B cells from the spleen were sorted with an EASYSEP MOUSE B-cell isolation kit (Stem Cell Technologies) and lysed using RIPA buffer (Santa Cruz) supplemented with a protease inhibitor (Orthovanadate, PMSF, anti-protease cocktail). Proteins were quantified with a Pierce BCA Protein Assay kit (Thermo Scientific) and denatured for 5 min at 95 °C. TGX Stain-free FastCast 12% acrylamide gels (Bio-Rad Laboratories) were used to separate proteins that were transferred to Transblot Turbo polyvinylidene fluoride membranes (Bio-Rad Laboratories) with a Transblot Turbo Transfer System. After blocking incubation with phosphate-buffered saline (PBS)-5% skim milk, the membrane was incubated with SATB1 primary antibody overnight at 4 °C. The membrane was incubated with secondary antibody in PBS-3% milk. Proteins were detected and quantified with PIERCE ECL Western blotting Substrate (Thermo Scientific) and ChemiDoc Touch Imaging System coupled to Image Lab J6.0 software (Bio-Rad Laboratories). The antibodies and concentrations used are listed in Supplementary Table S2. Unprocessed raw data are provided in Supplementary Fig. S1A.)
Enzyme-linked immunosorbent assay (ELISA)
Sera were collected from the blood of two-month-old mice. Supernatants were obtained after sorting B cells from spleens and in vitro LPS activation for 4 days. Plates were coated overnight with 1 µg/ml of primary antibody goat anti-mouse unlabeled IgM, IgG3, IgG1 or IgA (Southern Biotech) diluted in sodium carbonate buffer. Plates were blocked with PBS-3% bovine serum albumin for 1 h at 37 °C. Supernatant or diluted sera were incubated for 2 h at 37 °C, and then secondary antibody conjugated with alkaline phosphatase (Southern Biotech) was incubated for 1 h at 37 °C. Enzymatic reactions were performed by adding SigmaFastTM p-nitrophenyl phosphate tablets. DO was measured at 405 nm on a Thermo Scientific MULTISKAN FC. The antibodies and concentrations used are listed in Supplementary Table S2.
Flow cytometry
To analyze B-cell populations, spleen and bone marrow were collected, crushed and filtered through a 40 µm Clearline Strainer (Fisherbrand); lymphocyte numbers were quantified in a Cell-Dyn Emerald counter (Abbott). Cell suspensions were labeled in FACS buffer (PBS buffer (Eurobio), 10% BSA (Stem Cell Technologies) and 2 mM EDTA) containing antibodies for 20 min at 4 °C and analyzed on a BD LSRFortessa SORP flow cytometer (BD Bioscience). For intracellular staining, cells were first stained for surface markers. After washing, the cells were treated with the Intraprep fixation/permeabilization kit (Beckman Coulter, A07803) following the manufacturer’s instructions. Mean fluorescence intensity values were normalized to WT values; antibodies used are provided in Supplementary Table S2. Gating strategies to evaluate the proportions and numbers of B-cell subsets are provided in Supplementary Fig. S1.
RNA extraction
Pre-B cells, splenic resting B cells, in vitro-activated cells with LPS for 2 or 4 days, plasma cells and GC B cells were lysed with TRIZOL Reagent (Ambion Life Technologies), and RNA extraction was performed following the recommendations of the DirectZol RNA microprep kit (ZymoResearch).
RT‒qPCR
One microgram of RNA from B-cell subset samples was treated with DNase I Amplification Grade (Invitrogen), and reverse transcription was performed following the High Capacity cDNA Reverse Transcription kit protocol (Applied Biosystems). Quantitative PCR was performed on 20 ng cDNA using either SensiFAST Probe Hi-ROX or SensiFAST SYBR Hi-ROX kits (BioLine) on a Quant Studio III (Applied BioSystem). IgH and Igk primary transcripts were quantified as previously described [38]. Satb1 transcripts were measured by TaqMan assays with Mm.PT.58.13287891 (IDT), Rag 1 and Rag 2 transcripts with Mm01270936_m1 and Mm00501300_m1 (ThermoFisher), respectively. Transcript quantification was carried out with normalization to Hprt (Mm.PT.58.32092191 - IDT) for resting and in vitro-stimulated cells and to B220 (Mm.PT.58.32086646 - IDT) for GC B cells. Specific primers designed to amplify membrane and secreted Igµ transcripts, as well as Sµ transcripts, are listed in Supplementary Table S1.
Repertoire
Pre-B cells from bone marrow were enriched using a mouse CD25 MicroBead Kit (Miltenyi Biotec). Library preparation was adapted from methods previously described [39, 40]. Transcripts were amplified by 5’RACE PCR using reverse primer hybridizing within the µ gene and Cap race primer carrying unique molecular identifiers (UMIs) [41]. ProtoScript® II (New England Biolabs, Ipswich, MA) was used for reverse transcription, and amplicons were obtained using Phusion® High Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) according to the manufacturer’s instructions. (Primers are listed in Supplementary Table S1). Illumina adapter and tag sequences were added by primer extension [42]. The resulting libraries were sequenced on an Illumina MiSeq sequencing system using MiSeq Reagent kit V3 600 cycles. Paired reads were merged as previously described [42], and UMIs were treated with MIGEC software. Repertoire analysis was performed using the IMGT/HIGHV-QUEST online tool (http://imgt.org/) [43].
In vitro stimulation
Splenic B cells isolated with the EASYSEP MOUSE B-cell isolation kit (Stem Cell Technologies) were cultured at 1 × 106 cells/mL in RPMI 1640 medium (Eurobio) supplemented with 10% fetal bovine serum (Dutscher), 2 mM glutamine (Eurobio), 1% Eagle’s nonessential amino acids (Eurobio), 50 U/ml penicillin‒streptomycin (Gibco), 1 mM sodium pyruvate (Eurobio), and 129 µM 2-β-mercaptoethanol (Sigma‒Aldrich) in the presence of 1 µg/ml lipopolysaccharide (LPS-Invivogen). An aliquot of cells was collected for analysis and sample preparation at Day 2 or 4 after stimulation. The same isolation protocol and cell culture reagents were used for anti-CD40 + IL4 in vitro stimulation by replacing LPS with 5 µg/ml anti-CD40 antibody (R&D Systems, listed in Supplementary Table S2) and 20 ng/mL recombinant mouse IL4 (Peprotech). An aliquot of cells was collected for analysis and sample preparation at Days 3 and 4 after stimulation.
In vitro ethynil-deoxyuridine (EdU) incorporation
Splenic B cells were cultured as described above, and EdU was added 48 h later and incubated for 24 h. Cells were then processed by following the recommendations of the Click-it EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific). EdU incorporation was evaluated by flow cytometry on a BD LSRFortessa SORP flow cytometer (BD Bioscience).
NP-CGG immunization
Nitrophenylacetyl-Chicken Gamma Globulin (NP-CGG) immunization was realized by injecting 100 µg NP-CGG (N-5055C-5 Biosearch Technologies) precipitated with complete for the first intraperitoneal injection (Day 0) and incomplete Freund’s adjuvant for the second one at Day 12. Preimmune sera were collected before Day 0, and immunized sera were collected at Days 8 and 17. Mice were sacrificed on Day 17, and splenic GC B cells were sorted to extract DNA for SHM quantification by HTS.
Cell sorting
Plasma cells from spleen or plasmablasts from in vitro stimulation were sorted using B220 and CD138 surface markers on a BD FACS ARIA III (BD Bioscience). GC B cells were sorted using B220 and GL7 cell surface markers (Supplementary Table S2).
Somatic hypermutation analysis
SHM analysis was performed on B220+/GL7+ GG B cells sorted from Peyer’s patches or immunized spleens from wt, Satb1 cKO and Aicda KO mice. 5′Sµ, 3′JH4 and 3′Jκ5 intronic regions were amplified from 10,000 cells with specific primers (listed in Supplementary Table S1) using Phusion High-Fidelity DNA Polymerase (New England Biolabs) and according to the following program: denaturation (98 °C 10 s), hybridization (69 °C 30 s) and amplification (72 °C 1 min) for 38 cycles. Libraries were constructed with an Ion Xpress Plus gDNA Fragment Library kit (Cat. no. 4471269, Life Technologies) and sequenced on the Ion-Proton System S5. SHM frequencies were determined using raw data analyzed with the DeMinEr tool [37].
RNA sequencing
RNA-Seq analysis was performed on splenic resting B cells sorted with an Easysep Mouse B-cell isolation kit (Stem Cell Technologies), in vitro-activated B cells with LPS (bulk culture, at Day 2) and in vitro LPS-differentiated plasmablasts (sorted at Day 4). Sample quality controls and library preparation were performed at the GeT-Santé facility (Inserm, Toulouse, France, get.genotoul.fr). i/Quality control: RNA concentration and purity were determined using an ND-2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, USA). RNA integrity was checked with a Fragment Analyzer (Agilent Technologies, Santa Clara, USA) using the RNA Standard Sensitivity Kit. 260/280 purity ratios were all ≥1.8, and integrity indices were of good quality (8.3–10 RIN and >1.7 28 S/18 S ratios). ii/Library preparation (GeT-Santé): RNA-seq paired-end libraries were prepared according to Illumina’s protocol with some adjustments using the TruSeq Stranded Total RNA Gold Library Prep Kit (Illumina, San Diego, USA). Briefly, between 934–1000 ng of total RNA was first ribo-zero depleted using Illumina Ribo-Zero probes. Then, the remaining RNA was fragmented for 2 min and retrotranscribed to generate double-stranded cDNA. Compatible adaptors were ligated, allowing the barcoding of the samples with unique dual indices. Libraries were amplified by 12 PCR cycles, and an additional final purification step resulted in 280–700 bp fragments. Library quality was assessed using the HS NGS kit on the Fragment Analyzer (Agilent Technologies, Santa Clara, USA). iii/Library quantification (GeT-PlaGe): quantification and sequencing were performed at the GeT-PlaGe core facility (INRAE, Toulouse, France). Libraries were quantified by qPCR using the KAPA Library Quantification Kit (Roche, Basel, Switzerland) to obtain accurate quantification. iiii/Sequencing (GeT-PlaGe): libraries were equimolarly pooled, and RNA sequencing was then performed on one S4 lane of the Illumina NovaSeq 6000 instrument (Illumina, San Diego, USA) using the NovaSeq 6000 S4 v1.5 Reagent Kit (300 cycles), and a paired-end 2 × 150 bp strategy.
RNA-sequencing analysis
Paired-end reads were mapped on the GRCm38 mouse genome that was previously indexed with “Mus_musculus.GRCm38.dna.primary_assembly.fa” and “Mus_musculus.GRCm38.102.chr_patch_hapl_scaff.gtf” files from ENSEMBL release 102. Index and mapping steps were both performed with STAR v2.6.0c [44]. Then, featureCounts v2.0.1 [45] was used to count reads by gene. An R script named template_script_DESeq2_CL.r of SARTools [46] was run first with all count data to retrieve a PCA and check if biological variability was the main source of variance in the data. Then, the same script was run for each desired differential analysis with count data from defined reference and interest conditions. Differentially regulated genes with an adjusted p value of 0.05 and a fold change ≤−1.5 or ≥ 1.5 were selected for downstream analysis. Gene SYMBOLs were converted to ENTREZIDs with the bitr function of the R ClusterProfiler package [47]. The resulting ENTREZIDs and their associated log2fold change were then used to calculate enriched biological pathway profiles of different gene clusters (Down in cKO, Up in cKO, Down in WT and Up in WT) using the CompareCluster function of ClusterProfiler [47] with p value and q value thresholds set to 0.01 and 0.05, respectively. The resulting enriched functional profiles were filtered through a Gene Ontology list consisting of the hierarchical children of the following biological pathway terms: B-cell costimulation, B-cell selection, humoral immune response, immunoglobulin production, memory B-cell differentiation, regulation of the apoptotic process of mature B cells, and SHM (Supplementary Table S3). Terms that were enriched in all downregulated gene clusters or all upregulated clusters were discarded.
3C-HTGTS
3C-HTGTS was performed as previously described [48]. Briefly, 10 million resting and LPS-stimulated B cells (at Day 3) were crosslinked with 2% formaldehyde in 10% FCS PBS for 10 min at RT with rotation. Crosslinking was stopped by adding glycine at 0.1 M. Then, the cells were lysed in 50 mM Tris, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 1% TX-100 supplemented with protease inhibitor (ROCHE #11873580001). Nuclei were resuspended in 0.3% SDS for 1 h at 37 °C at 900 rpm and then neutralized with Triton TX-100 for 1 h. DNA restriction was performed using CviQ1 (Thermo Fisher ER0211) in B buffer (Thermo Fisher #BB5) overnight at 37 °C before heat inactivation for 25 min at 65 °C. Overnight ligation was performed at 16 °C and 300 rpm. Next, DNA was treated with proteinase K and RNase and cleaned with phenol/chloroform. After the 3 C step, the LAM-HTGTS protocol was performed [49]. Briefly, 3 C DNA was sonicated using the Bioruptor (Diagenode; two pulses at low intensity for 20 s), and 10 μg was used for the LAM-HTGTS step. Sµ bait was used for primer elongation. These single-stranded DNA fragments were incubated with streptavidin beads (Dynabeads C1 streptavidin beads; Invitrogen) overnight at RT and washed with BW buffer (1 M NaCl, 5 mM Tris-HCl pH 7.4, 0.5 mM EDTA pH 8.0). Universal I7 adaptors were ligated before nested PCR was performed with Sµ nested and universal I7 reverse primers (all primers are listed in Supplementary Table S1). After the Tagged PCR with I7 and I5 Illumina primers, PCR products were cleaned using a PCR clean-up kit (Macherey-Nagel REF#740609) and validated after migration on a BioAnalyser (Agilent). 3C-HTGTS libraries were sequenced on a 300 bp paired-end MiSeq V3 with 20% PhiX.
Analysis of 3C-HTGTS
Sequencing reads were aligned to the mm10 genome and processed as previously described [49]. Each 3C-HTGTS library plotted for comparison was normalized by randomly selecting a number of junctions equal to the total number of junctions present in the smallest library in the comparison set.
Results
Conditional deletion of Satb1 in murine B lineage cells
Since depletion of Satb1 in ES cells and mouse embryos resulted in multiple dysfunctions and did not allow animals to survive beyond 1 month of age [2], we evaluated the SATB1 contribution to B-cell development in a conditional knockout model inducing SATB1 depletion in B lineage cells from the pro-B-cell stage. In our model, initially derived from the Satb1tm1a allele, the Satb1 conditional allele (Satb1flx) contains exon 4 flanked with two LoxP sites that permit its specific deletion when coupled to one allele carrying a CRE recombinase insertion into the Cd79a gene (Cd79acre) [36] (Fig. 1A). Mice carrying the homozygous SATB1 deletion in B-lineage cells (Satb1flx/flx Cd79acre/+, referred as cKO conditional knockout) were compared to heterozygous littermates (Satb1flx/+ Cd79acre/+, referred as cHet: conditional heterozygous) and to wt littermates carrying an identical conditional allele but devoid of CRE expression (Satb1flx/+ Cd79a+/+, referred to as WT). Western blot experiments performed on sorted splenic B cells from these three genotypes confirmed that Satb1-exon4 deletion induced complete protein depletion (Fig. 1B, unprocessed raw data are provided in Supplementary Fig. S1).
Fig. 1.
A Conditional deletion of Satb1-exon 4 by Cd79acre/+ recombinase in B cells. Exon 4, cre site, frt site, conditional allele (Satb1flx) and wild-type allele (Satb1+) are indicated. B SATB1 protein quantification by Western blotting in splenic B cells from WT (Satb1flx/+), conditional heterozygous (Satb1flx/+ Cd79acre/+) and conditional homozygous (Satb1flx/flx Cd79acre/+) mice. C Satb1 transcript quantification by RT‒qPCR (normalized to Hprt) in Peyer’s patch naive and GC B cells sorted from WT mice (n = 8) from 3 independent experiments. D Bar graphs displaying absolute numbers of B-cell subsets determined by flow cytometry from bone marrow (per femur, n = 7–10) and spleen (n = 11–15) from 4–5 independent experiments. E Pie chart representation of IgHV family gene usage, quantified by RACE-RepSeq, expressed by pre-B-cell-enriched bone marrow populations in WT (n = 5) and cKO mice (n = 6) from 2 independent experiments. F Immunoglobulin serum isotype levels quantified by ELISA in 8- to 10-week-old WT and cKO mice (n = 8–15) from 4 independent experiments. Error bars represent SEM; p value was determined with two tailed Mann‒Whitney test; nonsignificant (ns) and significant differences are indicated (***p < 0.001)
SATB1 depletion in B lineage cells allows normal B-cell development and IgH repertoire
By querying the ImmGen database [50], we examined Satb1 expression in developing B-lineage cells and noted that all subtypes of B-lineage cells expressed Satb1 transcripts (Supplementary Fig. S2A). Common lymphoid progenitor and prepro B cells, also mentioned as Hardy fraction A, both displayed higher SATB1 expression. This was not surprising given that SATB1 has been reported to favor lymphocyte lineage differentiation from hematopoietic stem cells [23]. While a consistent drop in Satb1 transcription was observed at pro- and pre-B stages, a second wave of Satb1 gene expression occurred in transitional and mature resting B cells present in splenic follicles or the marginal zone as well as in splenic memory B cells. Satb1 gene expression further decreased in antigen-activated cells such as germinal center centrocytes to reach its minimal level in proliferating cell subsets such as centroblasts and plasmablasts (Supplementary Fig. S2A). In B cells from WT mice, we experimentally confirmed a significant decrease in Satb1 transcripts by RT‒qPCR assays when comparing naive and GC B cells sorted from Peyer’s patches (Fig. 1C). The same findings, corroborating expression profiles described in the ImmGen database, were very recently documented in a mouse model carrying a tomato-reporter transgene knocked into a Satb1 allele [35]. This converging evidence for variable expression of Satb1, according to B-cell subsets, supposes a narrow regulation and suggests an accurate function of this factor in the B-cell lineage.
To determine the SATB1 contribution throughout B-cell development and maturation, we analyzed B-cell populations from WT, cHet, and cKO mice by flow cytometry (Supplementary Fig. S1). Absolute numbers of B-lineage cell subsets from bone marrow (Fig. 1D left and middle) and spleen (Fig. 1D right) were not affected by Satb1 gene inactivation, suggesting, despite its contribution to lymphocyte lineage initiation [22], that SATB1 was not required for B-cell development. Our findings that early B-cell development was not impaired in our cKO mice excluded any function for SATB1 in B-lineage choice maintenance. These data were consistent with studies from Kanakura and Steidl groups [21, 22] that previously hypothesized that SATB1 function was restricted to stem cell renewal and fate of hematopoietic lineages.
Since SATB1 binding to the ASE region is known to modulate Rag gene expression in T cells and incidentally impact TCR rearrangements [33], the consistent expression of this factor in early committed and developing B cells could confer a similar function on Ig gene rearrangements. We investigated this point by first analyzing Rag gene expression by RT‒qPCR and found that sorted bone marrow pre-B cells from WT and cKO animals expressed similar transcript levels (Supplementary Fig. S2B). The strikingly different impact of SATB1 deletion on Rag expression in B and T cells could be indirectly explored by comparing DNA accessibility in the region close to Rag gene regions from WT B- and T-lineage cell precursors. Such a detailed study was provided by the consortium of the Immunological Genome Project that performed ATAC-seq in B and T-cell precursors [51]. This study indicates that while the DNA region encompassing the SATB1-binding ASE regulatory element was accessible in T-lineage precursor cells, this was not the case in bone marrow cell subsets of the B-cell lineage (a comparison focused on Rag gene loci is provided in Supplementary Fig. S2C). Notably, B-lineage precursors display other accessible regulatory regions, such as the Erag element located close to Rag gene regions. It is then reasonable to suppose that the ASE region is not active in the B-cell lineage, which could potentially explain normal Rag gene transcription in SATB1-deficient B-cell precursors. Given that SATB1 acts as a chromatin loop organizer in T cells [5, 34, 52–54], we suspected a potential effect of its deletion on IgH V region accessibility in developing B cells. To assess this point, we examined VDJ recombination diversity by repertoire sequencing experiments on RNA samples (RepSeq) extracted from pre-B-cell-enriched bone marrow fractions. Our data displayed an equivalent broad distribution of each rearranged and expressed V gene (Fig. 1E and Supplementary Fig. S2D) in WT and cKO mice. A similar representation of VH family usage indicated that SATB1 deletion did not hamper mechanisms leading to a diversified IgH VDJ repertoire in developing B cells. VDJ junction analysis in the preimmune repertoire of WT and Satb1 cKO models revealed comparable lengths of CDR3 regions as well as a rather comparable distribution of P and N nucleotides (Supplementary Fig. S2E), suggesting that IgHV region assembly and end-joining occur normally in the absence of STAB1. Altogether, our results indicate that SATB1 is dispensable for establishing the IgH repertoire and does not affect Rag expression or influence IgH locus accessibility. Given that SATB1 deletion did not impair VDJ rearrangements and B-cell development, further studies could then be performed on peripheral B-cell subsets in this model without any bias. In line with this statement, we determined whether SATB1 deletion could impact Ig isotype production and secretion in mice. Sera from WT, cHet and cKO animals were collected at two months of age, and IgM, IgG1, IgG3, and IgA levels were quantified by ELISA. Homozygous and heterozygous KO mice displayed serum Ig levels comparable to WT for each isotype (Fig. 1F), suggesting that SATB1 does not influence global antibody production.
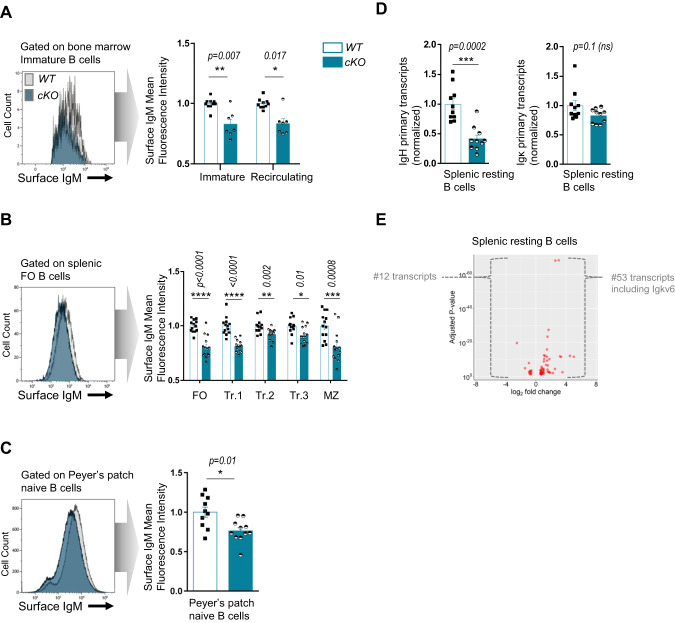
SATB1 depletion decreases IgH transcription in resting B cells
Despite normal bone marrow B-cell development and Ig secretion in cKO mice, we suspected that SATB1 depletion might influence IgH locus expression, given its capacity to bind MAR sequences in vitro [55] and to modulate MAR-containing reporter genes [29]. We first examined surface IgM expression levels, as a component of B-cell receptors, on immature and mature B-cell subsets from bone marrow and spleen compartments in our model. Upon measuring IgM mean fluorescence intensity (MFI) by flow cytometry, Satb1 cKO mice displayed consistent and significantly decreased IgM surface expression compared to WT mice on bone marrow immature and recirculating B cells (Fig. 2A) and on splenic transitional, marginal zone and follicular B-cell populations (Fig. 2B). The same significant decrease in IgM BCR expression, also observed on naive B cells sorted from Peyer’s patches (Fig. 2C), confirmed a specific function for SATB1 as a positive regulator of BCR expression in resting B cells. This role for SATB1 in BCR surface expression could explain its function as a contributing factor to B-cell survival, as described recently by Ozawa et al. [35]. Indeed, the knock-in reporter mouse model used in their study leads to inactivation of one Satb1 allele, and it is likely that such a deletion induces an intermediate surface IgM expression level similar to the one we observed in our heterozygous mice (Supplementary Fig. S3A). It is reasonable to suppose that reduced BCR expression on resting B cells reduces the ability of these cells to respond to anti-IgM-mediated in vitro stimulation. One fair interpretation of the survival defect described by Ozawa et al. [35] could be simply the consequence of reduced BCR expression in the knock-in model.
Fig. 2.
A Comparison of surface IgM fluorescence intensity, evaluated by flow cytometry, on immature and recirculating bone marrow B-cell subsets from two-month-old WT and homozygous cKO animals. One representative histogram is shown (left), and the bar graph (right) displays normalized surface IgM MFI in WT and cKO animals (n = 7–10 from 4 independent experiments). B Comparison of surface IgM fluorescence intensity, evaluated by flow cytometry, on follicular (FO), marginal zone (MZ), and transitional (Tr.1, Tr.2, Tr.3) splenic B-cell subsets from two-month-old WT and homozygous cKO animals. One representative histogram is shown (left), and the bar graph (right) displays normalized surface IgM MFI in WT and cKO animals (n = 13–14 from 5 independent experiments). C Comparison of surface IgM fluorescence intensity, evaluated by flow cytometry, on naive B cells sorted from Peyer’s Patches from two-month-old WT and homozygous cKO animals. One representative histogram is shown (left), and the bar graph (right) displays the normalized surface IgM MFI in WT and cKO animals (n = 10–11 from 3 independent experiments). D IgH and Igκ primary transcripts quantified by RT‒qPCR (normalized to Hprt) in splenic resting B cells sorted from WT and cKO mice (n = 6–10 from 5 independent experiments). E Volcano plot from RNA-seq indicating differentially expressed transcripts when comparing WT and cKO mouse (n = 2) resting B cells. Transcripts located within Ig gene loci are indicated. Error bars represent SEM; p value was determined by two tailed Mann‒Whitney test; nonsignificant (ns) and significant differences are indicated (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001)
The decreased BCR expression observed in SATB1-deficient B lymphocytes raised the question of a potential Ig gene transcription defect in our model. Quantification of IgH primary transcription by RT‒qPCR in resting splenic B cells showed a significant 2-fold decrease in transcripts running through the JH4 intron in cKO mice compared to WT mice (Fig. 2D). We also evaluated the respective proportion of membrane and secreted Igµ transcripts (Supplementary Fig. S3B). Our data showed that SATB1 deletion does not impair alternative splicing of the Igµ chain transcript. Although not statistically significant, a similar downward trend was also observed for Igκ transcription in mutants (Fig. 2D). MAR sequences flanking both sides of the IgH cEµ intronic enhancer are able to bind SATB1 [56]. Whereas binding of equivalent regions in the Igκ locus has never been shown, it has to be noted that only one upstream MAR is associated with the iEκ enhancer. The presence of either one or two MAR sequences surrounding these intronic enhancers could potentially explain the differences observed for IgH and Igκ transcription in our Satb1 KO model. It is tempting to speculate that, through its DNA binding in proximity to intronic enhancers, SATB1 could physiologically potentiate their transcriptional effect in resting B cells. However, this hypothesis is unlikely since IgM BCR expression was never compromised in resting B-cell populations of mice devoid of either Eµ, iEκ full regions or their associated-MARs [57–60]. In contrast, the literature reports that BCR expression on resting B cells was decreased upon deletion of components of the 3’ regulatory regions of both IgH [61] and Igκ [62] loci, arguing for the importance of close contact between 3’ enhancers and respective promoters of rearranged V exons. In resting B cells, such loops have been reported within the IgH locus in many studies [63]. In line with a proposed function for SATB1 as a promoter-enhancer loop regulator [54], a rational hypothesis could be that this MAR binding protein participates in physical interactions between rearranged VH exons and the 3’RR and consequently enhances Ig chain transcription in resting B cells. To evaluate such chromatin loop interactions, we performed 3C-HTGTS experiments using a bait within the Sµ region [48] of resting B cells from wt and mutant mice. Our data showed that loop interactions between Eµ and VH regions still occur at frequencies that do not significantly differ in resting B cells from both genotypes (Supplementary Fig. S4A); this suggests that SATB1 is not required to form chromatin loops in naive and LPS-activated B cells.
In line with the broad chromatin organization function proposed for SATB1 in T cells, we then performed total RNA-seq analysis of both WT and SATB1-deficient B-cell subsets. The resulting datasets were submitted to principal component analysis to validate reproducibility between samples (Supplementary Fig. S5A). When comparing datasets obtained from resting splenic B cells, our analysis only revealed 65 genes displaying significant changes in expression: 53 were overexpressed, and only 12 were downregulated (log2 FC > + /−1.5; Fig. 2E and Supplementary Table S4). Indeed, our data support the hypothesis of an ambivalent function for SATB1, displaying both positive and negative regulatory actions for gene expression in resting B cells. Our study indicates that SATB1 depletion does not induce drastic transcriptional changes in resting B cells. In contrast to the T lineage [7, 27, 64], our data suggest that the intrinsic function of SATB1 in B cells may be relatively focused. In this cell type, SATB1 would have a distinctly different role from that of a major genome organizer.
SATB1 depletion increases IgH transcription and Ig synthesis in activated B cells
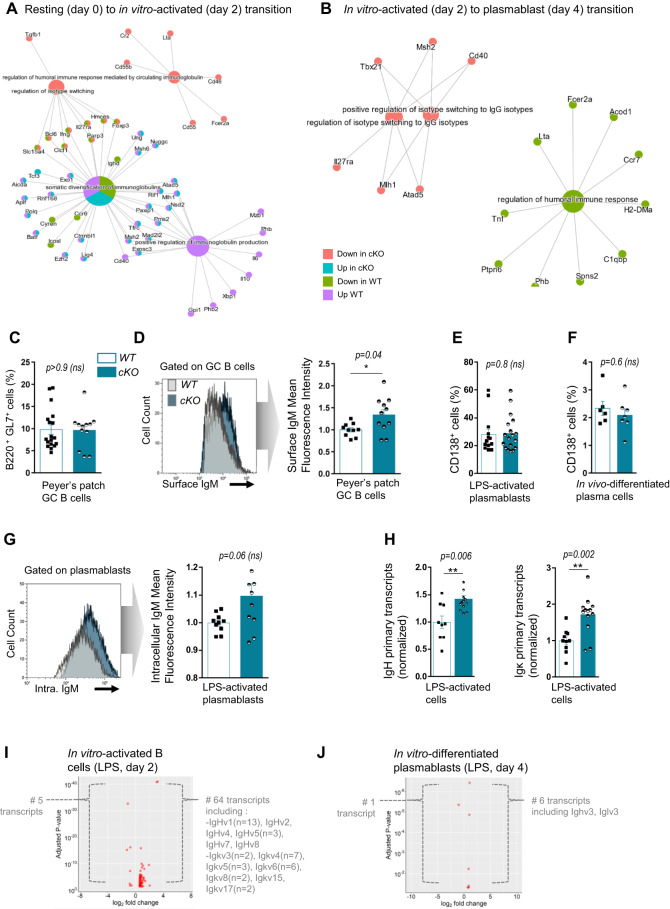
We sought to evaluate the effect of SATB1 depletion on B-cell activation in response to mitogenic and antigenic stimuli. We first performed in vitro stimulation of splenic B cells from both genotypes with LPS and carried out RNA-seq in a time-course manner on resting B cells (sorted at Day 0), activated B cells (bulk culture at Day 2) and plasmablasts (sorted at Day 4). Once sample reproducibility was validated (Supplementary Fig. S5A), we compared transcriptional programs induced by the transition from “resting to activated” and “activated to plasmablast” cell stages. Venn diagrams mainly indicate that both B-cell activation and differentiation programs induced by LPS were not drastically impaired upon SATB1 depletion. Indeed, following in vitro culture, a vast majority of transcripts displayed a common regulatory profile in WT and SATB1-deficient models, i.e., 5460 transcripts during the “resting to activated” transition and 2758 during the “activated to plasmablast” transition (Supplementary Fig. S5B). Nevertheless, we submitted differentially expressed genes from both transition programs to a gene ontology analysis for biological processes relevant to B-cell activation (see list in Supplementary Table S3). During the “resting to activated” transition, SATB1 depletion decreased the expression of a few genes involved in humoral immune responses mediated by circulating immunoglobulins (Fig. 3A, red dots), including Lta, a gene recently described as a SATB1 target in T cells [54], as well as Tgfb1 encoding TGFβ, the cytokine involved in the regulation of isotype switching to IgA. Strikingly, the analysis specified that SATB1 depletion increased Tcf3 expression in activated B cells (Fig. 3A, blue dot), while its gene expression, encoding the E2A transcription factor, was unchanged in hematopoietic stem cells devoid of SATB1 [23]. During the “activated to plasmablast” transition, SATB1 depletion downregulates Cd40 and a few genes involved in the Ig class switching pathway (Fig. 3B, red dots); this effect on Cd40 targeting was expected since it was already observed upon SATB1 deletion in hematopoietic stem cells and T cells [23, 54].
Fig. 3.
A Visual representation of Gene Ontology enrichment analysis of genes significantly associated with biological pathways upon B-cell transition from resting (Day 0) to in vitro-activated (Day 2) subsets in WT and cKO mice. Datasets from RNA-seq experiments were used for the analysis. B Same representation as above for B-cell transition from in vitro-activated (Day 2) to plasmablast (Day 4) subsets. C Germinal center (GC) B-cell subsets evaluated by flow cytometry in Peyer’s patches of WT and cKO animals (n = 12–18 from 4 independent experiments). D Comparison of surface IgM fluorescence intensity evaluated by flow cytometry on Peyer’s patch GC B cells from two-month-old WT and homozygous cKO animals. One representative histogram is shown (left), and the bar graph (right) displays normalized surface IgM MFI in WT and cKO animals (n = 10–11 from 3 independent experiments). E Proportion of plasmablasts evaluated by flow cytometry, obtained following in vitro LPS activation (Day 4) of splenic B cells from WT and cKO animals (n = 13–18 from 6 independent experiments) (left). F Proportion of plasma cells in the spleen evaluated by flow cytometry following NP-CGG immunization of WT and cKO animals (n = 6–7 from 1 experiment). G Comparison of intracellular IgM fluorescence intensity evaluated by flow cytometry in vitro-differentiated plasmablasts (Day 4) from WT and homozygous cKO animals. One representative histogram is shown (left), and the bar graph (right) displays normalized intracellular IgM MFI in WT and cKO animals (n = 9–10 from 4 independent experiments). H IgH and Igκ primary transcripts quantified by RT‒qPCR (normalized to Hprt) in in vitro LPS-activated B-lineage cells (Day 4) of WT and cKO animals (n = 9–13 from 5 independent experiments). I Volcano plots from RNA-seq indicating differentially expressed transcripts when comparing WT and cKO mice (n = 2) in in vitro LPS-activated B cells (Day 2). Transcripts located within Ig gene V regions are indicated. J Same analysis as above in vitro LPS-differentiated plasmablasts (Day 4). Error bars represent SEM; p value was determined by two tailed Mann‒Whitney test; nonsignificant (ns) and significant differences are indicated (*p < 0.05; **p < 0.01)
In parallel, in such in vitro stimulation assays, B cells were tested for intrinsic abilities to proliferate, to undergo LPS-driven class switch recombination and to differentiate into plasmablasts. When measured by EdU incorporation, the ability of SATB1-deficient B cells to proliferate was similar to that of WT cells (Supplementary Fig. S6A). This suggested that while decreasing IgM BCR expression on naive cells, SATB1 deficiency is dispensable for cell cycle entry of splenic B cells upon activation of TLR pathways. The same normal proliferation capacity in response to TLR or CD40 triggering was also recently reported for B cells devoid of the entire Satb1 gene region [35]. In our model, normal in vitro proliferation observed in B cells devoid of SATB1 was consistent with the normal proportion of spontaneous GC B cells in Peyer’s patches of KO animals (Fig. 3C) or the equivalent proportions of splenic GC centroblasts and centrocytes obtained after NP-CGG immunization (Supplementary Fig. S6B). More strikingly, in clear contrast with the decreased IgH expression in resting B cells, such in vivo-activated cells express higher BCR levels, as evidenced by IgM surface labeling (Fig. 3D). This highlights that the ambivalent function for SATB1 clearly applies to Ig gene expression in developing mature B cells.
Class switching to IgG3 was examined by surface labeling of in vitro-activated splenic B cells by LPS at Day 4. The data showed equivalent numbers of IgG3-expressing cells in both WT and SATB1 KO models (Supplementary Fig. S6C), indicating that SATB1 is not required for IgG3 class switching. RNA-seq datasets from LPS-activated cells at Day 2 also provided relevant portrayals of the germline transcription landscape in the IgH locus of WT and SATB1-deficient models (Supplementary Fig. S6D). When examining germline transcription within the Iµ-donor and Iγ3- or Iγ2b-acceptor regions, both WT and SATB1-deficient models displayed identical profiles, suggesting that, in agreement with normal CSR to IgG3 observed at Day 4, SATB1 depletion does not impede LPS-induced germline transcription occurring prior to CSR.
When measured in the same in vitro assays, the intrinsic ability of splenic B cells to differentiate into plasmablasts (CD138+) upon LPS activation was also similar in WT and our SATB1 KO model (Fig. 3E). In agreement with the in vitro data, NP-CGG immunization of mice induced the generation of plasma cells in normal proportions in the absence of SATB1 (Fig. 3F). Altogether, these data suggest that SATB1 is dispensable for efficient differentiation into antibody-secreting cells.
However, once differentiated in vitro into plasmablasts, SATB1-deficient cells displayed higher levels of intracellular IgM when analyzed by flow cytometry (Fig. 3G). This increase in Ig production by plasmablasts was correlated with a significant increase in both IgH and Igκ primary transcripts (1.4 and 1.7, respectively) in LPS-activated cells from SATB1-deficient animals (Fig. 3H).
The effect of SATB1 deletion on global transcription of both IgH and IgL chains was also confirmed by comparing RNA-seq data from splenic B cells activated by LPS at Day 2 (Fig. 3J) or from sorted plasmablasts at Day 4 (Fig. 3K). In the first case, among the 64 genes found to be upregulated in SATB1-deficient models (log2 FC > + /−1.5), 20 corresponded to IgV H chain products, and 23 others were identified as IgV κL chain products (Fig. 3J and Supplementary Table S5). When comparing plasmablasts, IgV H chain and IgV λL chain products were strongly increased upon SATB1 depletion (Fig. 3K and Supplementary Table S6). Our data indicate that, beyond its modest effect on B-cell activation programs induced by LPS in vitro, the absence of SATB1 induces a more pronounced transcription of Ig genes. Moreover, STAB1 is dispensable for proliferation, CSR and differentiation into plasma cells.
In parallel to LPS stimulation, we performed in vitro stimulation with anti-CD40 + IL4 in our model to evaluate, in another activation context, cell differentiation and Ig intracellular content in the absence of SATB1. Our data show, as in LPS stimulation, that SATB1-deficient B cells differentiate efficiently into plasmablasts in response to the anti-CD40 + IL4 cocktail (Supplementary Fig S6E left). Notably, plasmablasts devoid of SATB1 also contained high levels of intracellular IgM, although upregulation was not, in this case, increased significantly (Supplementary Fig S6E right). However, our data unveil a switch in the regulatory function of SATB1 for Ig chain transcription, from positive in naive to negative in activated B cells. This striking effect persists until the terminal stage since SATB1-deficient plasma cells display a high content of Ig chains. However, SATB1 depletion does not allow plasma cells to produce more Ig since our deficient mice display broadly normal levels of serum antibody isotypes.
SATB1 depletion increases somatic hypermutation
Since our group recently reported a critical function of cEµ-associated MARs for SHM of the IgH locus in mouse models [65], and according to historic studies in the Igκ locus [59], it was questionable whether MAR binding proteins, such as SATB1, could be involved in targeting somatic mutations of Ig genes. We first evaluated the ability of SATB1-deficient B cells to support somatic mutations by analyzing global SHM within intronic regions, not subject to antigen selection, located immediately downstream from V exons of both IgH and Igκ loci and upstream from IgH Sµ regions in spontaneous GC B cells from Peyer’s patches. As a possible regulatory mechanism, transcription taking place in the corresponding regions targeted by AID was also quantified in this subset. Second, to more accurately evaluate the ability of SATB1-deficient B cells to undergo SHM in response to specific antigens, we immunized mice with NP-CGG [66]. By analogy to B cells devoid of MARsEµ regions, in which SHM machinery gains access more frequently to the region downstream from cEµ (upstream from Sµ) [67], we carefully quantified SHM in this same region (Fig. 4A, scheme). In GC B cells sorted from Peyer’s patches, while Sµ transcription was not significantly increased (Fig. 4A, left bar graph, p = 0.3), we found a significant increase in mutations in the absence of SATB1 (1.88 mut/Kb) compared to WT SATB1 (1.44 mut/Kb) (Fig. 4A, middle bar graph, p = 0.02 and Supplementary Table S7A). A similar and significant increase in SHM in this region was also observed in splenic GC B cells sorted after NP-CGG immunization: while WT cells barely reached 0.22 mut/Kb, SATB1-deficient cells underwent twofold more mutations, reaching 0.41 mut/Kb (Fig. 4A, right bar graph, p = 0.03 and Supplementary Table S7B). This common feature shared by MARsEµ KO- and Satb1 KO-B cells suggests that SATB1 could be involved in limiting access of the SHM machinery to donor Sµ regions in cells undergoing SHM. When transcription and SHM were quantified downstream from the rearranged IgH variable regions (Fig. 4B, scheme) in GC B cells sorted from Peyer’s patches, homozygous Satb1 deletion increased transcription (Fig. 4B, left bar graph, p = 0.01) downstream from JH4 and raised, although not significantly, SHM frequencies (Fig. 4B, middle bar graph, p = 0.05 and Supplementary Table S7C). These findings were clearly confirmed upon NP-CGG immunization. Since the B-cell response to NP-CGG challenge is preferentially dominated by mutated clones expressing the VH186.2 segment [66], quantification of base substitutions in the JH4 intron downstream from the VH186.2-rearranged exons is considered a reliable hallmark of antigen-induced SHM. Indeed, in SATB1-deficient GC B cells from immunized mice, NP-CGG-induced mutations were significantly increased within the JH4 intron: mutant cells displayed 5.06 mut/Kb, while WT cells only reached 3.28 mut/Kb (Fig. 4B, right bar graph, p = 0.02 and Supplementary Table S7D). When, in the intronic region downstream from Jκ5 of the Igκ locus (Fig. 4C, scheme), global transcription was significantly increased in mutant samples, (Fig. 4C, left bar graph, p = 0.03), both spontaneous and NP-CGG-induced GC B cells displayed an increased trend of mutation frequencies upon SATB1 depletion (Fig. 4C, middle and right bar graphs and Supplementary Table S7E, F). Since B-cell responses to DNP hapten preferentially involve Ig composed of Igλ1 light chains [68], it is likely that changes in mutation frequency within the Igκ light chain loci, in this case not significant, probably underestimated any potential SHM increase induced by SATB1 depletion. While our Gene Ontology enrichment analysis detected the “Somatic diversification of Igs” as a differentially regulated pathway during the “resting to activated” transition, the expression of major actors of mismatch repair (MMR) and base excision repair (BER) pathways, such as Ung, Msh2, Msh6, Pms2, and Polq, was similarly upregulated in both wt and cKO samples (Fig. 3A). Beyond global pathway analysis, it was also necessary to verify in detail whether SATB1 depletion could affect, through its transcription factor function, the expression of any potential factors, including AID, involved in SHM. Indeed, when extracted from RNA-seq datasets, the expression of 16 genes involved in SHM was unchanged upon SATB1 deletion in both LPS-activated B cells and LPS-induced plasmablasts (Supplementary Fig. S7A). Within Ig gene regions submitted to SHM analysis, substitution frequencies calculated at each base displayed a globally increased pattern (although not significant) that did not offer any hypothesis regarding the origin of the changes (Supplementary Fig. S7B). It is well established that AID targeting SHM is coupled to transcription initiated at V promoters [69]. Given the significant increase in primary transcription occurring at IgH and Igκ in GC B cells devoid of SATB1 (Fig. 4B, C), one straightforward hypothesis to explain the global SHM increase in our model could be an overall increase in AID targeting of Ig genes. Interestingly, in line with this hypothesis, we recently highlighted increased AID deamination coupled to increased transcription in our mouse model devoid of MARsEµ regions when bred into a DNA repair-deficient background [67]. Surprisingly, while genomic deletion of MAREik or MARsEµ regions in the mouse decreased SHM [59, 67], suppression of SATB1 led to the opposite effect in the regions downstream from the rearranged variable segments of the IgH and Igκ loci. This finding also suggests a protective function for SATB1 against mutations. In this case, since the regions involved are located upstream from Eµ and Eiκ, one could propose that the potential protective effect of SATB1 could take place in resting B cells in which SHM is not expected to happen. Moreover, the discrepancy between the effect of MARsEµ deletion and SATB1 depletion on targeted mutations upstream from intronic enhancers suggests that other interacting factors, beyond SATB1, participate in the complex regulation of cis-acting MARs. While we recently proposed that MARsEµ favors error-prone repair in its upstream regions to optimize SHM [67], our current study suggests that SATB1 does not participate in the unfaithful repair processes associated with SHM.
Fig. 4.
Comparison of primary transcription and somatic hypermutation in WT and cKO animals, taking place in various regions of Ig genes targeted by AID (left schemas), in spontaneous GC B cells sorted from Peyer’s patches (n = 7–10 from 4 independent experiments, left and middle bar graphs). Somatic hypermutation in splenic GC B cells sorted after NP-CGG immunization (n = 7–8 from 4 independent experiments, right bar graphs). Data were obtained by NGS (Ion proton) combined with DeMinEr filtering, an accurate dedicated method to quantify AID-induced substitutions in B-lineage cells (50). A Transcription and SHM upstream from the IgH Sµ region. B Transcription and SHM within the IgH intron downstream from VJ558 (consensus) to JH4-rearranged exons (in spontaneous GC B cells) or in the same region downstream from V186.2 to JH4-rearranged exons (in NP-CGG-induced GC B cells). C Transcription and SHM within the Igk intron downstream from all-rearranged Jκ5 segments. Error bars represent SEM; p value was determined by two-tailed Mann‒Whitney test; nonsignificant (ns) and significant differences are indicated (*p < 0.05)
Taken together, our data suggest that SATB1 deletion increases the SHM of Ig genes through a transcription-coupled mechanism that probably favors AID targeting.
Discussion
The fact that SATB1 plays a major role as a “genome organizer” in hematopoietic- and T-lineage cells [7, 28, 30] suggested that it is also important in B-lineage cells, a cell type that also undergoes fine developmental regulation of its expression [35]. By using a conditional deletion model in B cells, our study fills in gaps concerning the function of SATB1 in this lymphocyte lineage. In contrast to its function in early-developing T cells, SATB1 is dispensable for the establishment of the Ig gene repertoire and overall early B-cell development in the bone marrow; this result is consistent with the pioneer SATB1-null mouse model in which B-cell populations could be observed in the spleen [2].
When looking at regions involved in Rag gene regulation, accessibility data suggest that, unlike its critical function in T-cell precursors, the ASE region is not active in B-lineage cells. This difference suggests that the Rag gene regulation process evolved separately in the B and T lineages, at least with regard to the regulatory function of SATB1. However, our model reveals that SATB1 is involved in the control of Ig gene transcription in mature B cells. Although previously unknown in the B lineage, our findings once again point to a dual function for SATB1 depending on the activation stage. We show that SATB1 promotes Ig gene transcription in resting B cells, while in activated B cells, it acts as a repressor. In contrast to the T-cell lineage, where SATB1 is considered a major regulatory factor of the enhancer/superenhancer network [53, 54], SATB1 deletion does not induce major disruptions in the B-cell transcriptome. Our invalidation model shows that only a reduced number of genes expressed during B differentiation are impacted by SATB1 deletion. In agreement with the repressive function of this factor in activated B cells, our study also shows that Ig genes are the predominant targets of SATB1 in activated B cells. Our 3C-HTGTS data indicate that SATB1 is not required to establish chromatin loops between Eµ and VH regions in resting and activated B cells. A rational hypothesis could be that the effect of SATB1 on IgH transcription is mainly due to its function as a transcription factor rather than a genome organizer. Given the critical effect of SATB1 depletion on Ig gene transcription, it would certainly be interesting to evaluate SATB1 physiological binding to regulatory regions in Ig gene loci in resting and activated B cells.
By clarifying that SATB1 is an essential activator of Ig chain transcription and consequently BCR expression in resting B cells, our study extends recent work published by Yokota’s team [35] and proposes a rational explanation for the survival function of SATB1 at this stage. These two studies also point out the importance of SATB1 expression levels, which could, as demonstrated in HSCs [22, 23], fine-tune some critical genes or regulatory pathways in lymphocytes. Beyond the physiological decrease in Satb1 expression between naive and activated B stages, it is possible that this gene undergoes splicing modifications in the B-cell lineage, as proposed in developing T cells by Spillianakis’ group [70]. The stability of the SATB1 protein, according to its transcript variants, might also be critical for its function in mature B-cell subsets, as already proposed in T-lineage cells by Galande’s group [19]. Interestingly, the dual function of SATB1 also observed in the B lineage, switching from activator to repressor of transcription in activated B cells, is reminiscent of the molecular switch observed upon phosphorylation and acetylation of this factor [16].
A surprising observation from our study is that SATB1 deletion increases somatic hypermutation of the IgH locus, extending into the Sµ region. It is very likely that the Igκ locus is similarly influenced. A direct impact of SATB1 on key factors involved in SHM, such as AID and proteins involved in BER and MMR pathways, can be excluded since normal induction of their transcripts was observed in the absence of SATB1. In this respect, SATB1 could play a protective role against SHM in resting cells. This finding should be read in conjunction with our recent study of the MAR regions associated with the Eµ enhancer of the IgH locus [67], which is known to bind SATB1 [1]. The SATB1-induced increase in SHM could logically be correlated with the observed increase in primary transcription of the variable regions of Ig H and L chains. However, it is not excluded that SATB1 modifies the accessibility of these regions to AID or its cofactors. Since it is proposed that SATB1 stabilizes the unpaired DNA regions against unwinding [13], such an action of SATB1 in the MARs-Eµ region could contribute to the protection of its surrounding regions against unwanted SHM. In line with the putative protective function of SATB1, it has been shown that this protein acts as an accessory factor of BER through its interaction with Oxo-guanine-glycosylase 1 (OGG1) [71], a DNA glycosylase that usually promotes error-free repair and is not involved in Ig gene SHM [72].
Further study of these modifications of SATB1 in B cells will be necessary to clarify the origin of its versatility. It has recently been proposed that SATB1 isoforms are subject to phase separation in T cells [70]. Consistent with its localization to PML nuclear bodies [73, 74], this mode of regulation deserves to be explored in the context of the B-cell nucleus. Moreover, since chromatin loop extrusion mechanisms are now proven to be critical for Ig gene regulation [75], it might be relevant to evaluate a potential implication of SATB1 in this process.
Supplementary information
unprocessed original images & Supplementary figures S1 to S7
Acknowledgements
The authors are grateful to the BISCEm unit (Univ. Limoges, UAR 2015 CNRS, US 42 Inserm, CHU Limoges) for technical support regarding DNA‒RNA sequencing, cytometry experiments and the animal core facility. We are grateful to Emeline Lhuillier and the Genotoul Plateau GeT-Santé facility (https://get.genotoul.fr) for technical assistance with RNA sequencing, to Mehdi Alizadeh from Etablissement Français du Sang (Rennes; France) for assistance with Repertoire sequencing and to Christelle Oblet for technical help with Western blotting. This work benefitted from data assembled by the ImmGen consortium [50]. MT, CB and OM were supported by PhD fellowships from the French Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation and the Fonds Européen de Développement Régional (FEDER). This work was supported by La Ligue Contre le Cancer (comités 87, 23 to EP and SLN); the Fondation ARC pour la recherche sur le cancer (PJA 20181207918 to EP and PhD continuation fellowship to MT and CB), Institut CARNOT CALYM, INCa-Cancéropôle GSO Emergence (to EP). We are grateful to Drs. Charalampos Spilianakis, Jeanne Moreau and Amélie Bonaud for critical reading of the manuscript, helpful comments and edits.
Author contributions
MT, CB, OM, SB, and CC performed the experiments. MT analyzed the data. EP and SLN conceived and supervised the study. MT and OM developed the experimental model. JP performed the bioinformatic analysis. MT, CB, EP, and SLN wrote the manuscript.
Data availability
Raw data from RNA-seq, SHM, Rep-Seq, and 3C-HTGTS have been deposited in the European Nucleotide Archive database under accession number PRJEB52320.
Competing interests
The authors declare no competing interests.
Contributor Information
Sandrine Le Noir, Email: sandrine.le-noir@unilim.fr.
Eric Pinaud, Email: eric.pinaud@unilim.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-023-01069-y.
References
- 1.Dickinson L, Tadashi J, Yoshinori K, Terumi K-S. A tissue-specific MARSAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–45. doi: 10.1016/0092-8674(92)90432-C. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–35. doi: 10.1101/gad.14.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai S, Han H-J, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expession regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- 4.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–5. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 5.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–88. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 6.Kohwi-Shigematsu T, Poterlowicz K, Ordinario E, Han H-J, Botchkarev VA, Kohwi Y. Genome organizing function of SATB1 in tumor progression. Semin Cancer Biol. 2013;23:72–9. doi: 10.1016/j.semcancer.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zelenka T, Spilianakis C. SATB1-mediated chromatin landscape in T cells. Nucl Austin Tex. 2020;11:117–31. doi: 10.1080/19491034.2020.1775037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Yang X, Chu X, Zhang J, Zhou H, Shen Y, et al. The structural basis for the oligomerization of the N-terminal domain of SATB1. Nucleic Acids Res. 2012;40:4193–202. doi: 10.1093/nar/gkr1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purbey PK, Singh S, Kumar PP, Mehta S, Ganesh KN, Mitra D, et al. PDZ domain-mediated dimerization and homeodomain-directed specificity are required for high-affinity DNA binding by SATB1. Nucleic Acids Res. 2008;36:2107–22. doi: 10.1093/nar/gkm1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamasaki K, Akiba T, Yamasaki T, Harata K. Structural basis for recognition of the matrix attachment region of DNA by transcription factor SATB1. Nucleic Acids Res. 2007;35:5073–84. doi: 10.1093/nar/gkm504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol. 1998;141:335–48. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo J, Lozano MM, Dudley JP. Nuclear matrix binding regulates SATB1-mediated transcriptional repression. J Biol Chem. 2005;280:24600–9. doi: 10.1074/jbc.M414076200. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh RP, Shi Q, Yang L, Reddick MP, Nikitina T, Zhurkin VB, et al. Satb1 integrates DNA binding site geometry and torsional stress to differentially target nucleosome-dense regions. Nat Commun. 2019;10. 10.1038/s41467-019-11118-8. [DOI] [PMC free article] [PubMed]
- 14.Kumar PP, Purbey PK, Ravi DS, Mitra D, Galande S. Displacement of SATB1-Bound histone deacetylase 1 corepressor by the human immunodeficiency virus type 1 transactivator induces expression of interleukin-2 and its receptor in T cells. Mol Cell Biol. 2005;25:1620–33. doi: 10.1128/MCB.25.5.1620-1633.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr Opin Genet Dev. 2007;17:408–14. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, et al. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231–43. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Purbey PK, Singh S, Notani D, Kumar PP, Limaye AS, Galande S. Acetylation-dependent interaction of SATB1 and CtBP1 mediates transcriptional repression by SATB1. Mol Cell Biol. 2009;29:1321–37. doi: 10.1128/MCB.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khare SP, Shetty A, Biradar R, Patta I, Chen ZJ, Sathe AV, et al. NF-κB signaling and IL-4 signaling regulate SATB1 expression via alternative promoter usage during Th2 differentiation. Front Immunol. 2019;10:667. doi: 10.3389/fimmu.2019.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patta I, Madhok A, Khare S, Gottimukkala KP, Verma A, Giri S, et al. Dynamic regulation of chromatin organizer SATB1 via TCR-induced alternative promoter switch during T-cell development. Nucleic Acids Res. 2020;48:5873–90. doi: 10.1093/nar/gkaa321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balamotis MA, Tamberg N, Woo YJ, Li J, Davy B, Kohwi-Shigematsu T, et al. Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol Cell Biol. 2012;32:333–47. doi: 10.1128/MCB.05917-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Will B, Vogler TO, Bartholdy B, Garrett-Bakelman F, Mayer J, Barreyro L, et al. Satb1 regulates the self-renewal of hematopoietic stem cells by promoting quiescence and repressing differentiation commitment. Nat Immunol. 2013;14:437–45. doi: 10.1038/ni.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi Y, Yokota T, Satoh Y, Okuzaki D, Tokunaga M, Ishibashi T, et al. Variable SATB1 levels regulate hematopoietic stem cell heterogeneity with distinct lineage fate. Cell Rep. 2018;23:3223–35. doi: 10.1016/j.celrep.2018.05.042. [DOI] [PubMed] [Google Scholar]
- 23.Satoh Y, Yokota T, Sudo T, Kondo M, Lai A, Kincade PW, et al. The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity. 2013;38:1105–15. doi: 10.1016/j.immuni.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen J, Huang S, Rogers H, Dickinson LA, Kohwi-Shigematsu T, Noguchi CT. SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood. 2005;105:3330–9. doi: 10.1182/blood-2004-08-2988. [DOI] [PubMed] [Google Scholar]
- 25.Hawkins SM, Kohwi-Shigematsu T, Skalnik DG. The matrix attachment region-binding protein SATB1 interacts with multiple elements within the gp91phox promoter and is down-regulated during myeloid differentiation. J Biol Chem. 2001;276:44472–80. doi: 10.1074/jbc.M104193200. [DOI] [PubMed] [Google Scholar]
- 26.Fujii Y, Kumatori A, Nakamura M. SATB1 makes a complex with p300 and represses gp91phox promoter activity. Microbiol Immunol. 2003;47:803–11. doi: 10.1111/j.1348-0421.2003.tb03438.x. [DOI] [PubMed] [Google Scholar]
- 27.Naik R, Galande S. SATB family chromatin organizers as master regulators of tumor progression. Oncogene. 2019;38:1989–2004. doi: 10.1038/s41388-018-0541-4. [DOI] [PubMed] [Google Scholar]
- 28.Papadogkonas G, Papamatheakis D-A, Spilianakis C. 3D genome organization as an epigenetic determinant of transcription regulation in T cells. Front Immunol. 2022;13:921375. doi: 10.3389/fimmu.2022.921375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohwi-Shigematsu T, Maass K, Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–10. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 30.Burute M, Gottimukkala K, Galande S. Chromatin organizer SATB1 is an important determinant of T-cell differentiation. Immunol Cell Biol. 2012;90:852–9. doi: 10.1038/icb.2012.28. [DOI] [PubMed] [Google Scholar]
- 31.Kakugawa K, Kojo S, Tanaka H, Seo W, Endo TA, Kitagawa Y, et al. Essential roles of SATB1 in specifying T lymphocyte subsets. Cell Rep. 2017;19:1176–88. doi: 10.1016/j.celrep.2017.04.038. [DOI] [PubMed] [Google Scholar]
- 32.Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18:173–83. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao B, Naik AK, Watanabe A, Tanaka H, Chen L, Richards HW, et al. An anti-silencer– and SATB1-dependent chromatin hub regulates Rag1 and Rag2 gene expression during thymocyte development. J Exp Med. 2015;212:809–24. doi: 10.1084/jem.20142207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng D, Li Z, Qin L, Hao B. The role of chromatin organizer Satb1 in shaping TCR repertoire in adult thymus. Genome. 2021;64:821–32. doi: 10.1139/gen-2020-0139. [DOI] [PubMed] [Google Scholar]
- 35.Ozawa T, Fujii K, Sudo T, Doi Y, Nakai R, Shingai Y, et al. Special AT-rich sequence-binding protein 1 supports survival and maturation of naive B cells stimulated by B cell receptors. J Immunol. 2022;208:1937–46. doi: 10.4049/jimmunol.2101097. [DOI] [PubMed] [Google Scholar]
- 36.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103:13789–94. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin OA, Garot A, Le Noir S, Aldigier J-C, Cogné M, Pinaud E, et al. Detecting rare AID-induced mutations in B-lineage oncogenes from high-throughput sequencing data using the detection of minor variants by error correction method. J Immunol. 2018;201:950–6. doi: 10.4049/jimmunol.1800203. [DOI] [PubMed] [Google Scholar]
- 38.Tinguely A, Chemin G, Péron S, Sirac C, Reynaud S, Cogné M, et al. Cross talk between immunoglobulin heavy-chain transcription and RNA surveillance during B cell development. Mol Cell Biol. 2012;32:107–17. doi: 10.1128/MCB.06138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascal V, Dupont M, de Rouault P, Rizzo D, Rossille D, Jeannet R, et al. Demultiplexing Ig repertoires by parallel mRNA/DNA sequencing shows major differential alterations in severe COVID-19. iScience. 2023;26:106260. doi: 10.1016/j.isci.2023.106260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouk C, Roland L, Gachard N, Poulain S, Oblet C, Rizzo D, et al. Continuous MYD88 activation is associated with expansion and then transformation of IgM differentiating plasma cells. Front Immunol. 2021;12:641692. doi: 10.3389/fimmu.2021.641692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turchaninova MA, Davydov A, Britanova OV, Shugay M, Bikos V, Egorov ES, et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat Protoc. 2016;11:1599–616. doi: 10.1038/nprot.2016.093. [DOI] [PubMed] [Google Scholar]
- 42.Javaugue V, Pascal V, Bender S, Nasraddine S, Dargelos M, Alizadeh M, et al. RNA-based immunoglobulin repertoire sequencing is a new tool for the management of monoclonal gammopathy of renal (kidney) significance. Kidney Int. 2022;101:331–7. doi: 10.1016/j.kint.2021.10.017. [DOI] [PubMed] [Google Scholar]
- 43.Boice M, Salloum D, Mourcin F, Sanghvi V, Amin R, Oricchio E, et al. Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell. 2016;167:405–18.e13. doi: 10.1016/j.cell.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 46.Varet H, Brillet-Guéguen L, Coppée J-Y, Dillies M-A. SARTools: a DESeq2- and EdgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Yoon HS, Chapdelaine-Williams AM, Kyritsis N, Alt FW. Physiological role of the 3’IgH CBEs super-anchor in antibody class switching. Proc Natl Acad Sci USA. 2021;118:e2024392118. doi: 10.1073/pnas.2024392118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat Protoc. 2016;11:853–71. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The Immunological Genome Project Consortium. Heng TSP, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, et al. The immunological genome project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–4. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, et al. The cis-regulatory atlas of the mouse immune system. Cell. 2019;176:897–912. doi: 10.1016/j.cell.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohwi-Shigematsu T, Kohwi Y, Takahashi K, Richards HW, Ayers SD, Han H-J, et al. SATB1-mediated functional packaging of chromatin into loops. Methods. 2012;58:243–54. doi: 10.1016/j.ymeth.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng D, Chen Y, Dai R, Bian S, Xue W, Zhu Y, et al. Chromatin organizer SATB1 controls the cell identity of CD4+ CD8+ double-positive thymocytes by regulating the activity of super-enhancers. Nat Commun. 2022;13:5554. doi: 10.1038/s41467-022-33333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zelenka T, Klonizakis A, Tsoukatou D, Papamatheakis D-A, Franzenburg S, Tzerpos P, et al. The 3D enhancer network of the developing T cell genome is shaped by SATB1. Nat Commun. 2022;13:6954. doi: 10.1038/s41467-022-34345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickinson LA, Dickinson CD, Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J Biol Chem. 1997;272:11463–70. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 56.Dickinson LA, Kohwi-Shigematsu T. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol Cell Biol. 1995;15:456–65. doi: 10.1128/MCB.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marquet M, Garot A, Bender S, Carrion C, Rouaud P, Lecardeur S, et al. The Eμ enhancer region influences H chain expression and B cell fate without impacting IgVH repertoire and immune response in vivo. J Immunol Balt Md 1950. 2014;193:1171–83. doi: 10.4049/jimmunol.1302868. [DOI] [PubMed] [Google Scholar]
- 58.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Ig kappa light chain intronic enhancer/matrix attachment region impairs but does not abolish V kappa J kappa rearrangement. Immunity. 1996;4:377–85. doi: 10.1016/S1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 59.Yi M, Wu P, Trevorrow KW, Claflin L, Garrard WT. Evidence that the Igkappa gene MAR regulates the probability of premature V-J joining and somatic hypermutation. J Immunol Balt Md 1950. 1999;162:6029–39. [PubMed] [Google Scholar]
- 60.Sakai E, Bottaro A, Davidson L, Sleckman BP, Alt FW. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc Natl Acad Sci USA. 1999;96:1526–31. doi: 10.1073/pnas.96.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garot A, Marquet M, Saintamand A, Bender S, Le Noir S, Rouaud P, et al. Sequential activation and distinct functions for distal and proximal modules within the IgH 3’ regulatory region. Proc Natl Acad Sci USA. 2016;113:1618–23. doi: 10.1073/pnas.1514090113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the kappa light chain intronic enhancer and 3’ enhancer in kappa rearrangement and demethylation. Nat Immunol. 2002;3:463–8. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 63.Bruzeau C, Moreau J, Le Noir S, Pinaud E. Panorama of stepwise involvement of the IgH 3’ regulatory region in murine B cells. Adv Immunol. 2021;149:95–114. doi: 10.1016/bs.ai.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 64.Yokota T, Kanakura Y. Role of tissue-specific AT-rich DNA sequence-binding proteins in lymphocyte differentiation. Int J Hematol. 2014;100:238–45. doi: 10.1007/s12185-014-1602-2. [DOI] [PubMed] [Google Scholar]
- 65.Martin OA, Thomas M, Marquet M, Bruzeau C, Garot A, Brousse M, et al. The IgH Eµ-MAR regions promote UNG-dependent error-prone repair to optimize somatic hypermutation. Front Immunol. 2023;14:1030813. doi: 10.3389/fimmu.2023.1030813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cumano A, Rajewsky K. Clonal recruitment and somatic mutation in the generation of immunological memory to the hapten NP. EMBO J. 1986;5:2459–68. doi: 10.1002/j.1460-2075.1986.tb04522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin O, Thomas M, Marquet M, Garot A, Brousse M, Bender S et al. The IgH Eμ -MAR regions promote UNG-dependent error-prone repair to optimize somatic hypermutation. Immunology. 2022. 10.1101/2022.08.15.503996. [DOI] [PMC free article] [PubMed]
- 68.Reth M, Hämmerling GJ, Rajewsky K. Analysis of the repertoire of anti-NP antibodies in C57BL/6 mice by cell fusion. I. Characterization of antibody families in the primary and hyperimmune response. Eur J Immunol. 1978;8:393–400. doi: 10.1002/eji.1830080605. [DOI] [PubMed] [Google Scholar]
- 69.Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–14. doi: 10.1016/S1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 70.Zelenka T, Tzerpos P, Panagopoulos G, Tsolis KC, Papamatheakis D-A, Papadakis VM et al. SATB1 undergoes isoform-specific phase transitions in T cells. Front. Cell Dev. Biol. 2022;11. 10.3389/fcell.2023.1242481. [DOI] [PMC free article] [PubMed]
- 71.Kaur S, Coulombe Y, Ramdzan ZM, Leduy L, Masson J-Y, Nepveu A. Special AT-rich sequence-binding Protein 1 (SATB1) functions as an accessory factor in base excision repair. J Biol Chem. 2016;291:22769–80. doi: 10.1074/jbc.M116.735696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winter DB, Phung QH, Zeng X, Seeberg E, Barnes DE, Lindahl T, et al. Normal somatic hypermutation of Ig genes in the absence of 8-hydroxyguanine-DNA glycosylase. J Immunol Balt Md 1950. 2003;170:5558–62. doi: 10.4049/jimmunol.170.11.5558. [DOI] [PubMed] [Google Scholar]
- 73.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, et al. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 74.Tan J-AT, Sun Y, Song J, Chen Y, Krontiris TG, Durrin LK. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J Biol Chem. 2008;283:18124–34. doi: 10.1074/jbc.M800512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Zhang X, Dai H-Q, Hu H, Alt FW. The role of chromatin loop extrusion in antibody diversification. Nat Rev Immunol. 2022;22:550–66. doi: 10.1038/s41577-022-00679-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
unprocessed original images & Supplementary figures S1 to S7
Data Availability Statement
Raw data from RNA-seq, SHM, Rep-Seq, and 3C-HTGTS have been deposited in the European Nucleotide Archive database under accession number PRJEB52320.