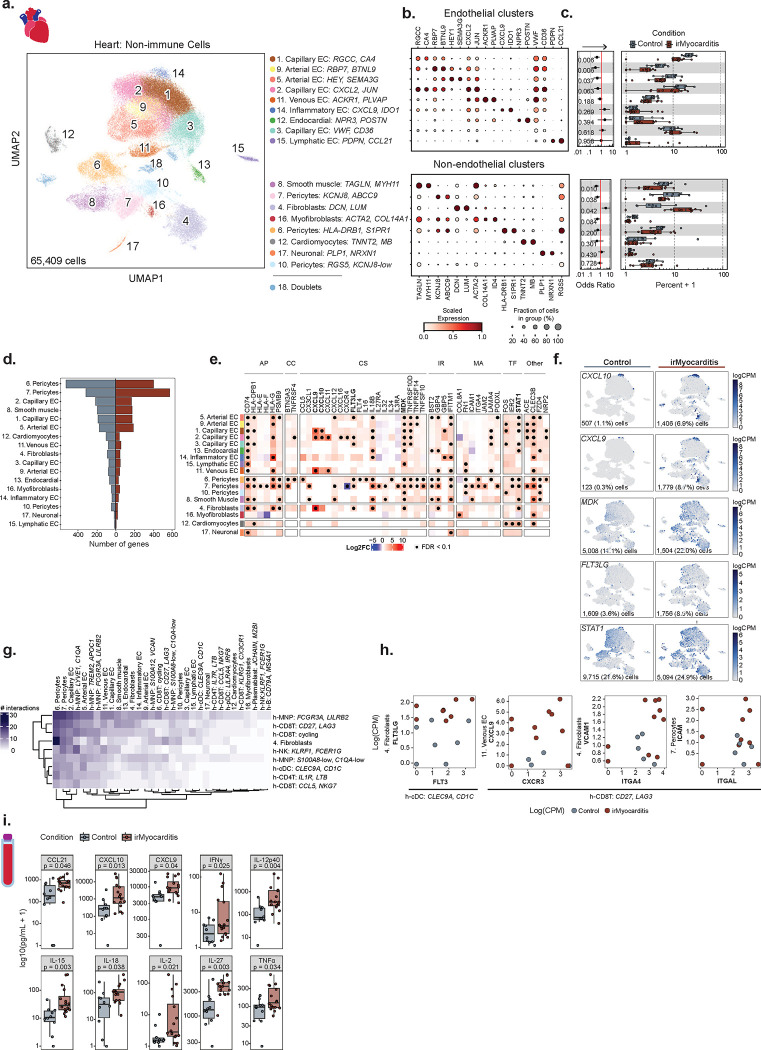
Figure 5. Immunomodulatory signals from non-immune intracardiac populations and circulating secreted factors nominate putative therapeutic targets for irMyocarditis.
a, UMAP embedding of 65,409 non-immune cells isolated from the heart, segregated by endothelial cells (top) and non-endothelial cells (bottom), and colored by the 18 defined cell subsets labeled on the right. Cell subset number was assigned according to the absolute number of cells detected per subset and are listed in order of ascending p-value for the associated analyses in panel c. b, Dot plot showing top marker genes for each non-immune subset. Dot size represents the percent of cells in the subset with non-zero expression of a given gene. Color indicates scaled expression across subsets. c, Cell subset differential abundance analysis comparing irMyocarditis cases (n = 12, red) to control (n = 8, blue). Left: logistic regression odds-ratio for association of non-immune cell subset abundance with irMyocarditis (OR > 1) versus control (OR < 1) for each subset. Unadjusted likelihood-ratio test p-values for each subset are shown. Right: boxplots show per patient intracardiac non-immune cell subset compositions, where each dot represents a patient. Composition is reported as the percent of cells from a patient in each subset. d, The number of differentially expressed genes (DEGs) for each cell subset when comparing irMyocarditis cases (right, red) to control (left, blue). e, Heatmap showing select DEGs (irMyocarditis versus control) grouped by the following biological themes: antigen presentation (AP), co-inhibition or co-stimulation (CC), cytokine signaling (CS), interferon response (IR), motility and adhesion (MA), transcription factors (TF), and “Other” genes of biological interest. Color scale indicates log2 fold-change difference between irMyocarditis cases and controls. Black dots indicate FDR < 0.1 (Wald test). f, Feature plots using color to indicate gene expression (logCPM) levels of the indicated genes expressed by control samples (left) or irMyocarditis samples (right) projected onto the non-immune cell UMAP embedding. Cell numbers and percentages represent gene expression across respective control or irMyocarditis non-immune cells. g, Heatmap reporting the number of significant interactions between intracardiac cell subsets as predicted by CellphoneDB. Significant CellphoneDB results were filtered for high-confidence interactions (see Methods). h, Scatter plots showing select predicted receptor:ligand pairs of interest from CellphoneDB analysis. Each point represents a donor (red = irMyocarditis, blue = control) and the x- and y-axes represent the logCPM of receptors and ligands, respectively, from the indicated cell subset. i, Serum concentrations of selected cytokines and chemokines in irMyocarditis cases and controls. In c, error bars represent 95% confidence intervals. In c and i, boxes represent the median (line) and interquartile range (IQR) with whiskers extending to the remainder of the distribution, no more than 1.5x IQR, with dots representing individual samples.