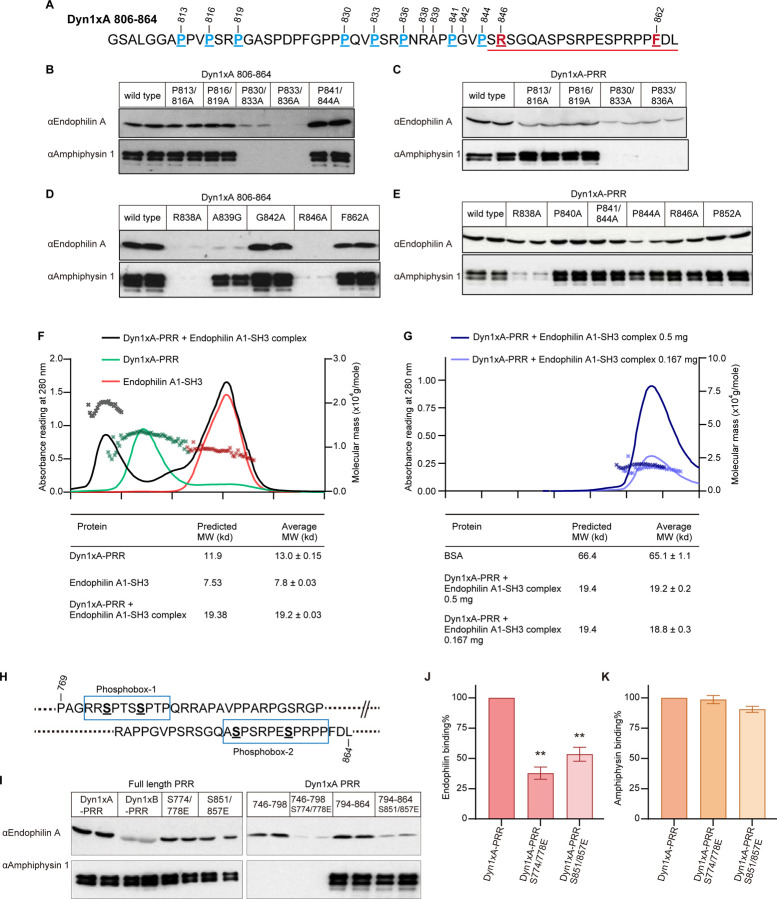
Figure 3. The interaction of Endophilin with both binding sites in Dyn1xA-PRR is phosphorylation-dependent in vitro.
(A) Amino acid sequence of Dyn1xA806–864. The amino acids for site-mutagenesis were underlined, and their positions marked with the number. The splice site after Ser845 in Dyn1xA-PRR is indicated with a red line.
(B-E) Samples from pull-down experiments from brain lysates with the indicated GST-tagged Dyn1xA806–864 or Dyn1xA-PRR and their specific point mutants, were run on gels, blotted and probed with antibodies to Endophilin A or Amphiphysin 1. Results shown are in duplicate samples from one of at least 3 independent experiments.
(F) SEC-MALS profiles for Dyn1xA alone (in green), Endophilin A SH3 alone (in red) and the complex of the two (in black) are plotted. The left axis is the corresponding UV (280 nm) signals, and the right axis shows the molar mass of each peak. The molecular weight of the complex was determined and tabulated in comparison with the predicted molecular weight.
(G) SEC-MALS profiles for a high concentration of Dyn1xA-PRR/Endophilin A SH3 complex (0.5 mg) (in dark blue) and a low concentration of Dyn1xA-PRR/endophilin A SH3 complex (0.167 mg) (in blue). The left axis is the corresponding UV (280 nm) signals, and the right axis shows the molar mass of each peak. The molecular weight of the complex was determined and tabulated in the table.
(H) A schematic diagram of phosphobox-1 and -2 in Dyn1xA-PRR each containing dual phosphorylated serines.
(I) Phosphomimetic mutants were made in the phosphoboxes. Samples from pull-down experiments with GST tagged PRD and both of its phosphomimetic mutants, S774/778E and S851/857E, GST-Dyn1xA 746–798 and Dyn1xA 746–798 S774/778E, GST-Dyn1xA 794–864 and Dyn1xA 794–864 S774/778E, were run on gels, blotted, and probed with antibodies to Endophilin A and Amphiphysin 1. Results are shown from one of three independent experiments.
(J, K) The amount of Endophilin A and Amphiphysin 1 bound to full-length Dyn1xA-PRR and its phosphomimetic mutants were quantified by densitometric analysis of the Western blots such as in panel I. n=2. All data were expressed as a percent of Dyn1xA-PRR Mean ± SEM. One-way ANOVA was applied (**p < 0.0001).