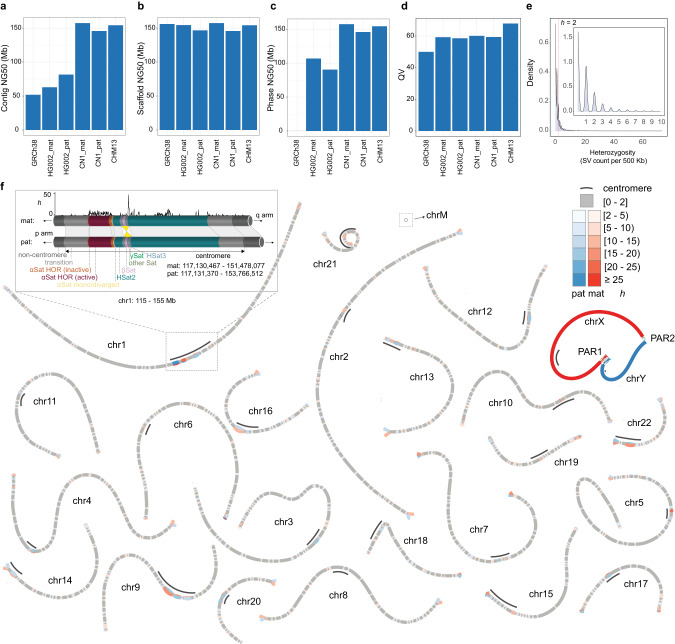
Fig. 1. Haplotype-resolved assembly of CN1 diploid genome.
a–d The contig NG50, scaffold NG50, phase NG50, and QV of GRCh38, HG002, CN1, and CHM13. e Whole-genome distribution of the heterozygosity rate (h). The heterozygosity rate is calculated as the SV count in each 500 kb window. f Visualization of the heterozygous regions between two haplotypes using bubbles. Here, h = 2 was set as the threshold for displaying bubbles. The homozygous regions are shown as single paths (grey), and the heterozygous regions at each heterozygosity rate are marked as bubbles. Regions with different h are shown in different shades. Centromeres (black lines) carry much higher h than other regions. The insert plot shows the structural variations in the centromere of chr1. αSat and HSat2 are the most divergent (window = 50 kb, step = 10 kb). An inversion, covering βSat and γSat, is shown between the paternal and maternal genomes (light yellow).