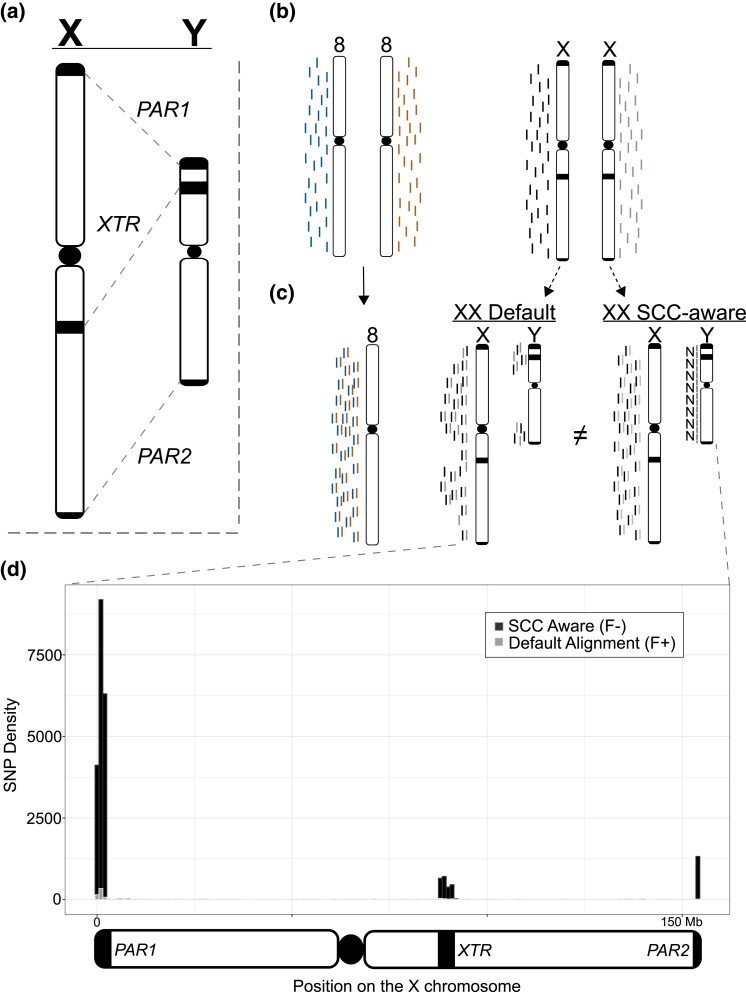
Fig. 1.
Overview of technical artifacts on sex chromosomes for read mapping and variant calling. a) Overview of regions of high-sequence identity between the X and the Y chromosomes. b) NGS reads originating from a karyotypically diploid XX individual. c) How reads from an XX-karyotype individual align to the Default reference and how masking the Y chromosome in these cases improves read MQ in these regions. d) Connecting changes in read mapping to differences in called variants across the X chromosome in the analysis presented in this paper. Dark/black regions can be viewed as presumed false negatives (variants missed with the Default reference), while light/gray areas can be viewed as false-positive calls (variants unique to the Default reference). Overlapping variant calls between the 2 reference genomes have been removed. The 3 regions that contain the most incorrect calls using the Default reference are the 2 PARs (beginning and end of the plot) and the XTR (just right of the plot center). SNPs are binned into 1 Mb windows.