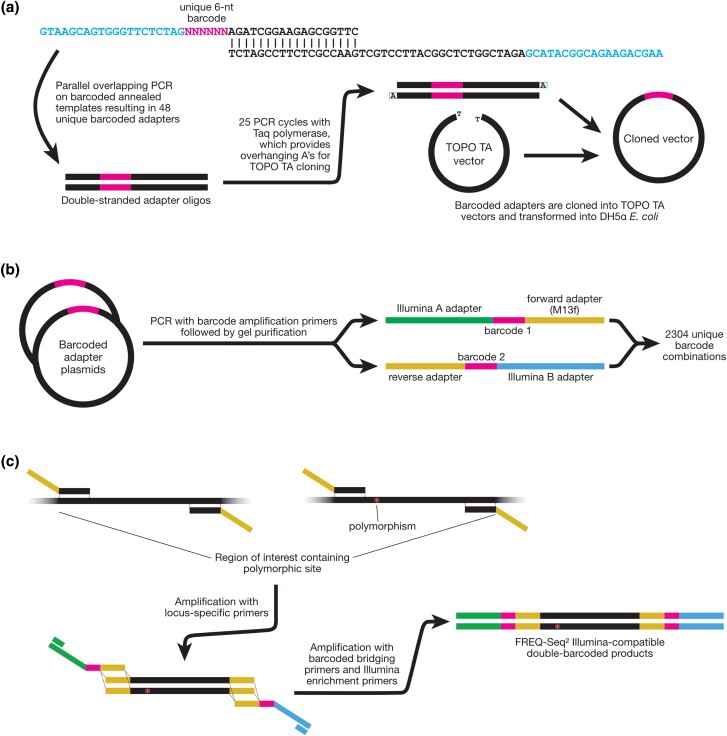
Fig. 1.
a) Protocol for generating the FREQ-Seq2 adapter library. Partially complementary single-stranded oligonucleotides containing the barcodes are annealed together, extended, and PCR amplified with primers corresponding to the regions in blue. Next, they are amplified with Taq polymerase to add overhanging adenosines, for cloning into the TOPO TA vector. After cloning into the plasmids, the vectors are transformed into competent DH5E. coli bacteria and plated, and plasmid DNA is extracted from the transformed bacteria. b) The Illumina-compatible FREQ-Seq2 barcoded bridging primers for paired-end sequencing can be amplified from the adapter plasmids using the same amplification primers used to generate the adapter fragments. These adapters can be used in conjunction with their corresponding FREQ-Seq barcoded adapters for double-barcoded labeling of fragment mixtures. c) To generate a FREQ-Seq2 sequencing library, amplification is first performed using locus-specific primers to produce a pool of fragments in a region of interest. These fragments contain adapters on each end that are complementary to the barcoded bridging primers, enabling double-barcoded labeling. Amplification is then performed using the barcoded bridging primers and enrichment primers, resulting in Illumina-compatible double-barcoded products.