Abstract
The ESKAPE pathogens are a group of bacteria that are a leading cause of health-care associated infections and are known to be agents of chronic, biofilm-mediated infections. These chronic bacterial infections often respond poorly to antibiotics and in some cases may require surgical intervention in order to cure the infection. As biofilms are often the critical mediator of a chronic infection, it is essential to develop therapies that target bacteria within the biofilm state. Herein, we report the development of a rapid, 96-well plate-based assay that employs conditions specific for each species to optimize biofilm production and allow for easy identification of differences in biofilm mass after treatment with anti-biofilm candidates. We used these ESKAPE-specific biofilm assays to test our previously identified Salmonella anti-biofilm small molecule compounds, JG-1 and M4, for anti-biofilm activity. The results demonstrated that JG-1 and M4 have anti-biofilm activity against Enterobacter spp., S. aureus, E. faecium, P. aeruginosa, and A. baumannii. In addition, we identified that M4 has significant antimicrobial activity against S. aureus and E. faecium at concentrations >10 μM (X μg/mL). These findings support the claim that JG-1 and M4 have broad-spectrum anti-biofilm activity, while M4 has antimicrobial activity against the Gram-positive members of the ESKAPE pathogens. Thus, these compounds have the potential to have a significant impact on treating multiple types of commonly encountered biofilm-mediated infections.
Keywords: ESKAPE pathogens, Antibiofilm small molecule compounds, Biofilm, Antibiotic
1. Introduction
Biofilms are communities of microorganisms, organized within a self-secreted extracellular matrix. Biofilms are ubiquitous in nature and contribute to an estimated 80% of human infections [1]. In clinical settings, biofilms are the primary growth state of bacteria and they can form both on abiotic surfaces, such as implantable medical devices, as well as directly on patient tissue [2]. Bacterial biofilm formation serves to adhere the cells to a surface as well as protect them from stressors in the environment. The extracellular polymeric substances (EPSs) produced by bacteria also serve as a layer of defense, protecting the bacteria from the host-defense response as well as certain classes of antibiotics. For example, the negatively charged alginate within the EPS of Pseudomonas aeruginosa can slow the diffusion of positively charged antibiotics such as aminoglycosides and prevent phagocytosis by leukocytes [3]. Within a biofilm state, bacteria are also signficiantly more resistant to antibiotics in general, being upwards of 1000-fold more resistant than their free-floating (planktonic) kin [4].
Biofilms are dynamic, demonstrating a great diversity of metabolic activity within different layers of its structure as well as the ability to respond to signals both from the environment and from each other. Biofilm formation occurs in stages, beginning with initial surface attachment, followed by maturation and then dispersal [2]. Planktonic cells first attach to a surface and then secrete EPS in order to adhere to each other and that surface. Depending on the environment, individual cells or aggregates of cells may disperse from the biofilm and attach to a new surface in order to establish a new nidus of infection. Many genes have been found to be differentially expressed in the various stages of biofilm development and may have the potential to be targets for biofilm prevention and disruption [[5], [6], [7], [8]]. Moreover, there is evidence that recently dispersed bacteria are more susceptible to antimicrobials than their non-biofilm or planktonic counterparts, suggesting a combination of anti-biofilm and antibiotic treatment may increase clearance of chronic infections [2,9,10].
The “ESKAPE” acronym refers to a group of bacteria including Enterobacter spp., Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterococcus faecium. Escherichia coli is often included in studies of the ESKAPE pathogens (then called ESKAPE-E), because it is also an emerging cause of serious disease in humans [11,12]. The ESKAPE pathogens are a major cause of biofilm-mediated infections and are estimated by the National Healthcare Safety Network to be involved in greater than 40% of infections in patients in the intensive care unit [13]. Antibiotic resistance is also prominent in the ESKAPE group. Biofilms can also accelerate dissemination of antibiotic resistance, as biofilm-based bacteria have increased rates of conjugation, increasing the rate of spread of plasmids harboring antibiotic resistance genes [14].
Given the importance of biofilms in the clinical setting, much research has been devoted to understanding the role of biofilms in chronic infection and identifying ways to prevent or treat them. To that end, we have previously identified two small molecules that can inhibit and disperse Salmonella biofilms in vitro and work cooperatively with ciprofloxacin in vivo in our mouse model of chronic gallbladder carriage [15]. Further, we have established that these compounds are non-toxic in multiple human cell lines and well-tolerated in mice [15]. Given the redundancy in biofilm promoters and components across many bacterial species, we hypothesized that these two compounds would also be efficacious in pathogens beyond Salmonella. In order to perform an initial screen, we chose to evaluate both compounds for activity against all the ESKAPE pathogens due to their diversity and relevance to chronic infections. There have been multiple studies that evaluate the efficacy of antibiofilm or antibiofilm/antibiotic combination therapies [16,17], but significant variations exist in the ESKAPE species used with a lack of uniformity in the methodology for biofilm establishment that makes interpreting and repeating results across many studies difficult [18]. Many of these in vitro methodologies require large volumes, involve difficult techniques, or restrict high-volume testing that prevents accommodation of large screens of potential anti-biofilm compounds against various ESKAPE pathogens. We present a description of a rapid attachment assay that can be used to test the anti-biofilm properties of compounds against the ESKAPE group.
2. Methods and materials
2.1. Bacterial strains, growth conditions, and compounds
Strains used include Enterobacter spp (Enterobacter cloacae subsp. cloacae (Jordan) Hormaeche and Edwards, subsp. nov. ATCC 13047), Staphylococcus aureus ATCC 29213, Staphylococcus aureus USA300, Klebsiella pneumoniae IA565, Klebsiella pneumoniae Top52, Klebsiella pneumoniae ATCC 13883, Acinetobacter baumannii ATCC 17978, Pseudomonas aeruginosa ATCC 27853, vancomycin-sensitive Enterococcus faecium, vancomycin-resistant Enterococcus faecium (acquired from the Ohio State University Hospital East Clinical Microbiology Lab), E. coli (acquired from Ohio State University Hospital East Clinical Microbiology Lab), E. coli MC4100, E. coli DH5α, E. coli (wild type enteroaggregative strain from Invitrogen).
Individual bacterial colonies were used to inoculate Luria Bertani broth (LB) (Enterobacter spp., K. pneumoniae, A. baumannii, P. aeruginosa) or tryptic soy broth (TSB) (S. aureus, E. faecium) for overnight (O/N) liquid cultures grown at 37 °C with aeration in a rolling drum.
Identification of compounds M4 and JG-1 were previously described (see Supplementary Fig. 1 for their respective structures) [15,19]. HCl-salted samples of M4 and JG-1 were stored as powders in −20 °C, shielded from light. Stock solutions were prepared in DMSO at a concentration of 100 mM and stored at −20 °C. All further dilutions were prepared in culture media such that the final DMSO concentration was no greater than 10% (v/v). In experiments where the effects of more than one concentration were tested, the concentration of the vehicle was standardized across all conditions by adding an equal volume of DMSO.
2.2. Media
Liquid media used include LB, TSB, M17 Broth (M17), and Brain Heart Infusion Broth (BHI). Biofilms were grown in either undiluted LB, LB diluted 1:20 in dH2O (1:20 LB), undiluted TSB, TSB diluted 1:20 in dH2O (1:20 TSB), TSB supplemented with 2.5 g/L dextrose (TSBG), TSB supplemented with 2.5 g/L dextrose and 3 mM FeSO4 (TSBGFS), TSB supplemented with 2.5 g/L dextrose and 150 μM hemin (TSBGH), undiluted M17, M17 supplemented with 3 mM FeSO4 (M17FS), or M17 supplemented with 150 μM hemin (M17H), or undiluted BHI.
2.3. Biofilm culture
2.3.1. Rapid attachment assay
Broadly, broth media (3 mL) was inoculated with a single isolated colony of the desired species and incubated at 37 °C overnight in order to allow the bacteria to reach log-phase growth. Overnight cultures were then normalized in media and diluted. A volume of 100 or 200 μL was pipetted into a standardized non-treated polystyrene 96-well plate. An un-inoculated media blank was included to use as a negative control and the plate was incubated for 24 h at either 30 or 37 °C.
2.3.2. Biofilm mass quantification
Biofilm mass was quantified using the crystal violet assay [15]. In brief, after incubation, the supernatant in each well was discarded and the wells washed by submerging the 96-well plate in dH2O and slowly swished. The wash was then repeated in fresh dH2O. The biofilms were then heat fixed at 60 °C for 1 h. A volume of crystal violet solution (6 mL phosphate buffered solution (PBS), 3.3 mL 1% (w/v) crystal violet solution, 333 μL isopropanol, 333 μL methanol) equal to the volume of culture previously added to wells was pipetted into each well and incubated at room temperature for 5 min. The crystal violet stain was discarded and the plates washed twice with dH2O. Any remaining water from the stained wells was removed and the remaining crystal violet bound to the biofilm was solubilized by adding an equal volume of 33% (v/v) acetic acid solution to each well. The OD570 of each well was then measured using a plate reader (Spectramax M3) in order to compare the biofilm mass present in the wells.
2.3.3. Biofilm inhibition and disruption assays
The optimal conditions for biofilm growth were used to determine the anti-biofilm properties of M4 and JG-1. The conditions used for each organism are described in Supplemental Table 1.
For inhibition assays, biofilms were grown as described above in the presence of either M4 or JG-1 (diluted in media from 100 mM stock solutions) or vehicle (DMSO) supplied in media at the time of inoculation. Biofilms were grown for the specified incubation time, temperature, and status prior to analysis via crystal violet staining as outlined in Supplemental Table 1.
For disruption assays, biofilms were initially grown in media alone as described above. Spent media was then removed and replaced with media containing the appropriate concentrations of anti-biofilm compounds (diluted from 100 mM stock solutions) or vehicle (DMSO). Biofilms were then incubated again as described prior to analysis via crystal violet staining.
2.3.4. IC50/EC50 determination
Half-maximal concentrations (the concentration of compound required to achieve a response halfway between baseline and maximum, defined here as 50% biofilm formed/remaining relative to the vehicle control) were calculated for both compounds in the inhibition assays (referred to as IC50 values) and disruption assays (referred to as EC50 values). IC50 values for biofilm inhibition were calculated using measurements of percent biofilm formed relative to vehicle (DMSO) after 24 h growth in the presence of various concentrations of JG-1 or M4 as described previously. EC50 values for disruption were calculated using measurements of percent biofilm remaining relative to vehicle after treating 24 h biofilms (grown as described previously) with various concentrations of JG-1 or M4 for an additional 24 h. Biofilms were quantified using the crystal violet assay as described previously.
2.3.5. Confocal microscopy
In order to visualize biofilm structures using confocal microscopy, biofilms of Enterobacter spp., K. pneumoniae, and P. aeruginosa were grown in 8-well chambered coverglasses (Thermo Fisher Scientific) by modifiying the microtiter biofilm inhibition and disruption assays described above.
Briefly for inhibition assays, Enterobacter spp. biofilms were grown as described above in the presence of either 5 μM M4, JG-1, or vehicle (DMSO) added to the media at the time of innoculation. Chambered coverglasses were incubated statically at 30 °C for 24 h. For disruption assays, Enterobacter spp. biofilms were grown in media without compound for 24 h statically at 30 °C. The spent media was then removed and replaced with media containing either 20 μM M4, 20 μM JG-1, or vehicle (DMSO) and reincubated for another 24 h.
For dispersal assays, K. pneumoniae biofilms were grown in a volume of 200 μL media without compound for 24 h nutating at 37 °C. The spent media was then removed and replaced with media containing either 80 μM M4, 80 μM JG-1, or vehicle (DMSO) and reincubated for another 24 h.
For inhibition assays, P. aeruginosa biofilms were grown as described above in the presence of either 20 μM M4, 20 μM JG-1, or vehicle (DMSO) added to the media at the time of innoculation. Chambered coverglasses were incubated nutating at 30 °C for 24 h. For disruption assays, P. aeruginosa biofilms were grown in media without compound for 24 h nutating at 30 °C. The spent media was then removed and replaced with media containing either 40 μM M4, JG-1, or vehicle (DMSO) and reincubated for another 24 h.
Following the above inhibition/disruption periods, the media was removed and the remaining biofilms were washed once with PBS. Bacterial cells and cellulose were labeled with the dyes SYTO9 (final concentration 5 μM; Molecular Probes) and calcofluor white (final concentration 30 μg/mL; Sigma-Aldrich) in 5% bovine serum albumin blocking buffer (5% BSA), respectively. After a 30 min incubation period at room temperature, the SYTO9 and calcofluor white mixture was removed. Subsequently, in order to visualize amyloid proteins (curli), the biofilm was incubated with human α-amyloid IgG (diluted 1:250 in 5% BSA; courtesy of Çagla Tükel, Temple University) for 30 min and then the primary antibody was removed and washed once with PBS. The biofilm was then incubated with Alexa Fluor 647 goat α-human IgG (1:1000 in 5% BSA; Invitrogen) for 30 min. The unbound secondary antibody was removed before fixing the biofilms with 4% paraformaldehyde solution (PFA) for 20 min prior to imaging. All incubations and washes were carried out at room temperature and chamber slides protected from direct light in order to prevent photobleaching.
Stained biofilms were visualized at 63X magnification using an inverted Zeiss LSM 800 confocal laser scanning microscope. Three-dimensional biofilm structures were imaged by capturing five representative Z-stacks per well for each treatment. For every slice within a Z-stack, the signal for each fluorophore was recorded separately: SYTO9-labeled cells were visualized at an excitation of 483 nm and an emission of 503 nm, calcofluor white-bound cellulose was visualized at an excitation of 365 nm and an emission of 435 nm, and Alexa Fluor 647-labeled amyloid was visualized at an excitation of 650 nm and an emission of 665 nm. Z-stacks were analyzed using Comstat2 software [20] in order to calculate values of biomass, average thickness, and maximum thickness for each individual biofilm component assessed.
2.3.6. Planktonic viability assay
To evaluate JG-1 and M4 for bactericidal or bacteriostatic capabilities, O/N liquid cultures of A. baumannii, P. aeruginosa, S. aureus, and E. faecium were grown in LB or TSB and were normalized to OD600 = 0.8 and diluted 1:1000 in LB or TSB containing JG-1, M4, or a vehicle control (DMSO). The effects of both low (10 μM) and high (80 μM) concentrations of JG-1 and M4 were assessed, and all conditions were normalized to a final DMSO concentration of 1% (v/v). Bacteria were incubated at 37 °C with aeration, and samples taken over the course of 24 h were enumerated by serial dilution and plating onto LB agar (incubated for 16 h at 37 °C).
3. Results
3.1. Biofilm assay development
Optimal conditions for biofilm production were determined for each species in the ESKAPE group as well as E. coli, and used in subsequent experiments (Supplemental Table 1). These assays were developed with the guidance of other previously published rapid attachment assays [21,22]. Utilizing optimal conditions for each organism allowed us to determine even subtle effects when testing the biofilm inhibitor compounds. Factors contributing to biofilm production, including optical density of the inoculum, liquid growth media, supplements to liquid growth media, and dilution of culture prior to addition to 96-well plates, were compared within each species in order to identify which combination of factors produced the most biofilm mass as measured by crystal violet staining. Multiple isolates of some species were compared for their biofilm producing capabilities in order to identify isolates that may be better suited for this rapid attachment assay. In determining the optimal conditions and isolates for each organism we prioritized conditions that produced the largest amount of biofilm as determined by crystal violet staining. However in some instances, these conditions presented other challenges that led us to designate an alternate set of conditions that improve the usability and/or reproducibility of the biofilm assay.
3.1.1. Enterobacter spp.
Overnight cultures of an isolate of Enterobacter spp. were grown in LB at 37 °C. The overnight culture was either diluted 1:10 in LB or 1:20 LB or normalized to OD600 = 0.1 or 0.8 prior to 1:10 dilution in LB, 1:20 LB, TSB, or 1:20 TSB. The diluted cultures were pipetted into 96-well plates and incubated statically for 24 h at 30 °C. The greatest biofilm masses were detected in Enterobacter spp. biofilms grown in 1:20 LB where the initial inoculum was normalized to OD600= 0.1. (Fig. 1A.). These biofilms were not statistically more significant than any other conditions (with the exception of LB- OD600=0.8) but represented the conditions with the largest mean difference between conditions tested (Supplemental Fig. 3A.).
Fig. 1.
Comparison of conditions tested to form optimal biofilms. (A–G.) Liquid cultures of isolates of E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, Enterobacter spp., and E. coli. were normalized to an optical density at 600 nm (OD600) between 0.1 and 0.8 and diluted in LB, TSB or BHI supplemented with or without dextrose (G), hemin (H), and/or FeSO4 (FS) in order to induce biofilm growth in 96-well plates. After incubation for 24 h, biofilm quantity was determined by staining with crystal violet (CV) and measuring the absorbance of CV at 570 nm (OD570). Bars represent the average and standard error of the mean of all replicates adjusted for background absorbance of the media used. N = 2–7 biological repeats each demonstrating similar relative differences between strains and conditions. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1.2. Staphylococcus aureus
Overnight cultures of isolates USA300 (MRSA) and ATCC 29213 (MSSA) were grown in TSB at 37 °C. The overnight cultures were normalized in either TSB or TSBG to OD600 = 0.8 and then further diluted 1:100. The diluted cultures were pipetted into 96-well plates and incubated statically for 24 h at 37 °C. MRSA and MSSA produced the largest biofilm mass when diluted and grown in TSB supplemented with dextrose (Fig. 1B.).
3.1.3. Klebsiella pneumoniae
K. pneumoniae strains ATCC 13883, Top52, and IA565 were compared for their biofilm producing capabilities. Overnight cultures of ATCC 13883, Top52, and IA565 were grown in LB at 37 °C and either diluted 1:1001 in LB, TSB, or BHI or normalized to OD600 = 0.8 and then further diluted 1:1001. The diluted culture was pipetted into 96-well plates and incubated for 24 h nutating at 37 °C. The largest biofilm masses were observed in K. pneumoniae biofilms grown in LB where the initial inoculum was not normalized (LB- OD600 = None), with Top52 and IA565 producing much more biofilm than ATCC 13883 (Fig. 1C.). Between the conditions tested for Top 52 and IA565, the LB- OD600 = None, strain IA565 had the narrowest confidence intervals (Supplemental Fig. 3B.).
3.1.4. Acinetobacter baumannii
Overnight cultures of A. baumannii strain ATCC 17978 were grown in LB at 37 °C. The overnight culture was diluted 1:100 in LB, 1:20 LB, TSB or 1:20 TSB or normalized to OD600 = 0.1 or 0.8 and further diluted 1:100 in LB, 1:20 LB, TSB, or 1:20 TSB. The diluted cultures were pipetted into 96-well plates and incubated statically for 24 h at 30 °C. The greatest biofilm masses were detected in A. baumannii biofilms grown in TSB where the initial inoculum was normalized to OD600 = 0.8. (Fig. 1D.). However, the high amount of crystal violet staining in this condition is close to the limit of detection of our instrument. In order to avoid the need to manipulate the protocol, we chose a slightly less effective, but more consistently measurable, set of conditions, listed in Supplemental Table 1, to use for future A. baumannii biofilm assays.
3.1.5. Pseudomonas aeruginosa
Overnight cultures of P. aeruginosa strain ATCC 27853 were grown in TSB at 37 °C. The overnight culture was normalized OD600 = 0.8 and further diluted 1:100 in LB, 1:20 LB, TSB or 1:20 TSB. The diluted cultures were pipetted into 96-well plates and incubated nutating for 24 h at 30 °C. P. aeruginosa biofilms grown in 1:20 LB produced the most biofilm mass (Fig. 1E.).
3.1.6. Enterococcus faecium
Overnight cultures of a vancomycin-sensitive E. faecium isolate (referred to as VSE) and two vancomycin-resistant (VRE) clinical isolates of E. faecium, referred to as CI#1 and CI#2, were grown in TSB at 37 °C. The overnight culture was either diluted 1:25 in TSB or TSBG or normalized to OD600 = 0.1–0.8 and then diluted 1:25 in TSB, TSBG, TSBGH, TSBGFS, M17, M17H, or M17FS. The diluted cultures were pipetted into 96-well plates and incubated statically for 24 h at 37 °C with 5% CO2. The largest biofilm masses were detected in E. faecium biofilms grown in either TSB supplemented with dextrose and hemin or FeSO4 and M17 supplemented with hemin or FeSO4. There was some variation in the optimal combination of media and supplements with the CI#2 strain, which produced the greatest amount of biofilm in TSBGH, and CI#1, which produced the greatest amount of biofilm in M17FS. Media without supplementation or only supplemented with dextrose was not sufficient to support appreciable growth of biofilms for any of the isolates. Regardless of media used, normalization of the initial inoculum to OD600 = 0.7 produced the most biofilm between the isolates (Fig. 1F.).
3.1.7. Escherichia coli
E. coli isolates were compared for their biofilm producing capabilities and optimal conditions for biofilm production. Overnight cultures of strains MC4100, DH5α, 2 clinical isolates of E. coli (CI#1 and CI#2), and an enteroaggregative strain (EA) were grown in TSB or LB at 37 °C. Overnight cultures of each were either normalized to OD600 = 0.8 and then diluted 1:100 in LB, 1:20 LB, TSB, or TSBG or were diluted without normalization 1:100 in TSB, TSBG, LB, or 1:20 LB. The diluted culture was added to 96-well plates and incubated statically for 24 h at 37 °C. There was great variability in biofilm mass produced by each isolate. Overall, the clinical isolates produced the largest biofilms with the greatest biofilm mass when grown in TSB and normalized to OD600 = 0.8. The DH5α strain performed the best when not normalized and grown in TSBG. The enteroaggregative strain and MC4100 produced little to no biofilm in all of the conditions tested (Fig. 1G.).
3.2. Compound inhibition and dispersion activity
3.2.1. Inhibition
Using the established optimal biofilm conditions for each organism, we tested JG-1 and M4 for anti-biofilm activity. To test the compounds, they were administered at the same time as the biofilm was initiated. This allowed us to identify the efficacy of the compound of preventing bacterial cells from forming a biofilm in otherwise optimal biofilm-promoting conditions. After incubation, the biofilm mass of wells treated with JG-1 or M4 (0.141–300 μM) were determined using the crystal violet assay. The percent of biofilm formed relative to the vehicle was calculated by comparing the vehicle biofilm mass to the JG-1 and M4 treated biofilm mass (Fig. 2A-B.). When cells were treated with JG-1, we observed a dose-dependent reduction in biofilm formation for Enterobacter spp., A. baumannii, and P. aeruginosa. When cells were treated with M4, we observed a dose-dependent reduction in percent biofilm formed in Enterobacter spp., S. aureus (MSSA and MRSA), A. baumannii, and E. faecium (VSE and CI#1). M4 more effectively inhibited biofilm formation of Enterobacter spp. than JG-1 [JG-1 IC50 = 8.06 μM (95% CI, 7.08–9.17 μM) and M4 IC50 = 0.80 μM (95% CI, 0.68–0.94 μM)] (Fig. 2E-F). Conversely, JG-1 more effectively inhibited biofilm formation of A. baumannii than M4 [JG-1 IC50 = 54.34 μM (95% CI, 37.92–94.25 μM) and M4 IC50 = 892 μM (95% CI, 0.36 μM - 2.19 M)] (Fig. 2E-F.). JG-1 effectively inhibited biofilm formation of P. aeruginosa [JG-1 IC50 = 18.75 μM (95% CI, 10.17–43.58 μM] (Fig. 2E.). M4 effectively inhibited the biofilm formation of MSSA [M4 IC50 = 9.38 μM (95% CI, 6.40–13.75 μM)] and MRSA, [M4 IC50 = 10.68 μM (95% CI, 7.88–14.48 μM] (Fig. 2F.). Additionally, M4 also effectively inhibited the biofilm formation of E. faecium, both the VSE isolate [M4 IC50 = 18.71 μM (95% CI, 2.05e−133-1.71e123 M)] and the clinical isolate [M4 IC50 = 8.72 μM (95% CI, 3.97–19.17 μM)] (Fig. 2F.). It was also noted that we observed an increase in biofilm formed when K. pneumoniae cells were treated with [M4]≥36 μM).
Fig. 2.
Biofilm/Planktonic cell inhibition after treatment with lead compounds. Evaluation of (A) JG-1 and (B) M4 efficacy in inhibiting biofilm formation and their effect on planktonic cells. Biofilms were grown in the presence of varying concentrations of compound or vehicle (DMSO) in 96-well plates for 24 h. The biofilm quantity formed was determined and compared to the vehicle. (C) Examining the effect of JG-1 and (D) M4 on planktonic cell growth. At the end of incubation, prior to evaluation of biofilm quantity, the OD600 of planktonic cells in each well was measured and the OD600 of wells with compounds were compared to the vehicle to identify the proportion of planktonic cells remaining after treatment with compound. Points represent the average and standard error of the mean of all replicates. (E) and (F) IC50 values corresponding to the inhibition of biofilm mass measured in A and B were calculated for JG-1 and/or M4 for Enterobacter. spp [JG-1 8.06 μM (95% CI, 7.08–9.17 μM) and M4 0.82 μM (95% CI, 0.68–0.94 μM)]), MSSA [M4 = 9.38 μM (95% CI, 6.40–13.75 μM)], MRSA, [M4 = 10.65 μM (95% CI, 7.84–14.48 μM], E. faecium (VSE) [M4 = 18.70 μM (95% CI, 1.93e−06- 1810 μM)], E. faecium CI#1 [M4 =8.71 μM (95% CI, 3.97–19.17 μM)], A. baumannii [JG-1 = 54.34 μM (95% CI, 37.92–94.25 μM) and M4 = 892 μM (95% CI, 0.37–2175 μM)], and P. aeruginosa [JG-1 = 18.75 μM (95% CI, 10.17–43.58 μM)]. IC50 values were calculated using GraphPad Prism 9.0 to plot normalized compound activity (percent biofilm formed) as a function of log10 drug concentration and fitting the dose response curve (log[inhibitor] vs. normalized response,variable slope). N = 2–6 biological repeats each demonstrating similar relative differences between strains and conditions.
To determine if compounds were inhibiting biofilm formation through a microbicidal or non-microbicidal mechanism, we evaluated the impact each compound had on planktonic cell growth. We noted that JG-1 reduced planktonic cells in the supernatant in assays with A. baumannii ([JG-1] ≥ 18 μM) and P. aeruginosa ([JG-1] ≥ 10 μM), while M4 showed reduction of planktonic S. aureus cells ([M4] ≥ 5 μM) (Fig. 2C-D.). This suggested that our reduction in biofilm formation in these species may be due to antibacterial activity. In addition, there was an increase in planktonic cells remaining after treatment of Enterobacter spp. with [M4] ≥ 18 μM (Fig. 2D.). There was no decrease in planktonic cell quantity in Enterobacter spp., S. aureus, K. pneumoniae, or E. faecium cells treated with JG-1 (Fig. 2C.). Similarly there was no change in planktonic cell mass in K. pneumoniae, A. baumannii, or P. aeruginosa treated with M4 and S. aureus and E. faecium cells treated with low concentrations of M4 (Fig. 2D.). This suggested that our results seen in Enterobacter spp. are specific to the biofilm phenotype and not a result of antibacterial activity. However, antibacterial activity may be responsible for the effects of JG-1 on A. baumannii and P. aeruginosa. Similarly, antibacterial activity may be partially responsible for the biofilm reduction in S. aureus and E. faecium when exposed to M4 at concentrations ≥5 μM.
3.2.2. Dispersion
In order to identify the biofilm dispersion properties of each compound, we administered JG-1 and M4 to biofilms that had been established over 24 h. This allowed us to evaluate the efficacy of our compounds in disrupting a pre-formed biofilm in otherwise biofilm-promoting conditions. 96-well plates were inoculated with each organism and allowed to incubate for 24 h in their respective biofilm-promoting conditions. After 24 h, the supernatant was replaced with media containing JG-1 or M4 (0.141–300 μM) or equal amounts of DMSO diluted in liquid media and allowed to incubate for an additional 24 h. The resulting wells were subsequently washed and the biofilm mass of each well was determined using the crystal violet assay (Fig. 3A-B.). We observed a dose-dependent reduction in biofilm mass in Enterobacter spp. biofilms treated with both compounds. M4 more efficiently reduced biofilm mass than JG-1 [M4 EC50 = 8.59 μM (95% CI, 6.01–12.95 μM) and JG-1 EC50 = 38.8 μM (95% CI, 28.1–57.2 μM)] (Fig. 3E.). We did not observe any effect on biofilms of S. aureus, K. pneumoniae, A. baumannii, or E. faecium when treated with JG-1 or the biofilms of A. baumannii or E. faecium treated with M4. Using the full range of concentrations of JG-1 tested we are unable to calculate an EC50 for P. aeruginosa despite the observed decrease in biofilm mass at concentrations >10 μM (Fig. 3A). However, if analysis is limited to [JG-1] >10 μM we can conclude that JG-1 effectively disperses P. aeruginosa biofilms [JG-1 EC50 = 123.1 μM (95% CI, 55.98 μM–270.8 μM)] Notably, we did observe that M4 treatment resulted in a slight increase in biofilm formation in S. aureus ([M4] ≥ 40 μM), K. pneumoniae ([M4] ≥ 72 μM), and P. aeruginosa ([M4] ≥ 10 μM). Similarly to what we observed in the inhibition assays, we noted that JG-1 reduced planktonic cells in the supernatant for A. baumannii ([JG-1] ≥ 18 μM) and P. aeruginosa ([JG-1] ≥ 10 μM), while M4 showed reduction of planktonic E. faecium (VRE) cells ([M4] ≥ 36 μM) (Fig. 3C-D.). In addition, there was an increase in planktonic cells remaining after treatment of Enterobacter spp. with [M4] ≥ 3 μM (Fig. 3D.).
Fig. 3.
Biofilm disruption after treatment with lead compounds. Evaluation of (A) JG-1 and (B) M4 efficacy in disrupting preformed biofilms of the ESKAPE pathogens. Biofilms were grown in 96-well plates for 24 h before being exposed to varying concentrations of compound for an additional 24 h. The amount of biofilm mass remaining after compound treatment was quantitated and compared to vehicle in order to determine the change in proportion of biofilm remaining. Points represent the average and standard error of the mean of all replicates. (C) Examining the effect of JG-1 and (D) M4 on planktonic cell growth. At the end of incubation with compound, prior to evaluation of biofilm quantity, the OD600 of planktonic cells in each well was measured and the OD600 of wells with compounds were compared to the vehicle to identify the proportion of planktonic cells remaining after treatment with compound. Points represent the average and standard error of the mean of all replicates. (E) EC50 values corresponding to the dispersion of biofilm mass measured in A and B were calculated for JG-1 and M4 for Enterobacter spp. [JG-1 = 37.25 μM (95% CI, 27.66–53.31 μM) and M4 = 9.32 μM (95% CI, 6.89–13.04 μM)]. EC50 values were calculated using GraphPad Prism 9.0 to plot normalized compound activity (percent biofilm remaining) as a function of log10 drug concentration and fitting the dose response curve (log[inhibitor] vs. normalized response, variable slope). Points represent the average and standard error of the mean of all replicates. N = 2–5 biological repeats, each demonstrating similar relative differences between strains and conditions.
3.2.3. Changes in biofilm structure
In addition to measuring changes in biofilm biomass after JG-1/M4 treatment, we used confocal microscopy to visualize the biofilm structure of selected compound-bacteria combinations. We specifically stained the biofilm cells and the extracellular matrix components cellulose and amyloid (curli), and quantified the biomass and thickness of each component individually. This experiement provides context for how the compound effects observed in the crystal violet assays translate into changes in biofilm structure.
To detect changes in biofilm structure associated with inhibitory activity of the lead compounds, normalized cultures of Enterobacter spp. and P. aeruginosa were incubated with JG-1, M4, or DMSO for 24 h prior to labeling for cellulose, amyloid, and bacterial cells (Fig. 4A and E.). We observed a significant decrease in the biomass, maximum thickness, and average thickness of amyloid and bacterial cells in the biofilms formed by Enterobacter spp. cells treated with compound in comparison to vehicle (Fig. 4B-D.). We also observed a decrease in cellulose in compound treated cells, although this was not statistically significant (Fig. 4B-D.). P. aeruginosa cells treated with JG-1 showed a non-significant decrease in biofilm biomass, maximum thickness, and average thickness of biomass of amyloid and cells (Fig. 4F-H.). P. aeruginosa cells treated with M4 showed a non-significant increase in biofilm biomass and maximum thickness of amyloid, and a significant increase in average biomass thickness of amyloid (Fig. 4F-G.). Additionally the M4 treated P. aeruginosa cells showed a non-significant increase in maximum thickess of biofilm cells (Fig. 4H.). We did not observe any significant difference in cellulose abundance in cells treated with either compound. These results were generally consistent with the observed biofilm inhibitory effect of JG-1 and M4 on Enterobacter spp. and JG-1 on P. aeruginosa.
Fig. 4.
Evaluation of changes in biofilm structure associated with JG-1/M4 inhibitory activity. (A.-D) Enterobacter spp. and (E-H) P. aeruginosa biofilms were grown in the presence of 5 μM or 20 μM of compound or vehicle (DMSO), respectively, in 8-well chambered coverglasses for 24 h. (A and E) Biofilms were stained with SYTO9, calcofluor white, and a human α-amyloid antibody in order to visualize cells (green), cellulose (blue), and curli fimbriae/amyloid (red), respectively, prior to fixation in 4% PFA. Representative z-stacks (N =5 per treatment) were captured at 63X magnification using a Zeiss LSM 800 laser scanning confocal microscope. (B-D and F-H) COMSTAT2 software was used to calculate the (B and F) biomass, (C and G) biofilm maximum thickness, and (D and H) biomass average thickness of biofilm z-stacks for each individual component (cells, cellulose, and amyloid). Data are presented as the mean +/- SD. Annotations above the data denote values that are significantly different from the vehicle control (DMSO), as determined by two-way ANOVA with Dunnett’s test for multiple comparisons; * = p < 0.05, ** = p < 0.01, *** = p<0.001, ns= not significant (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
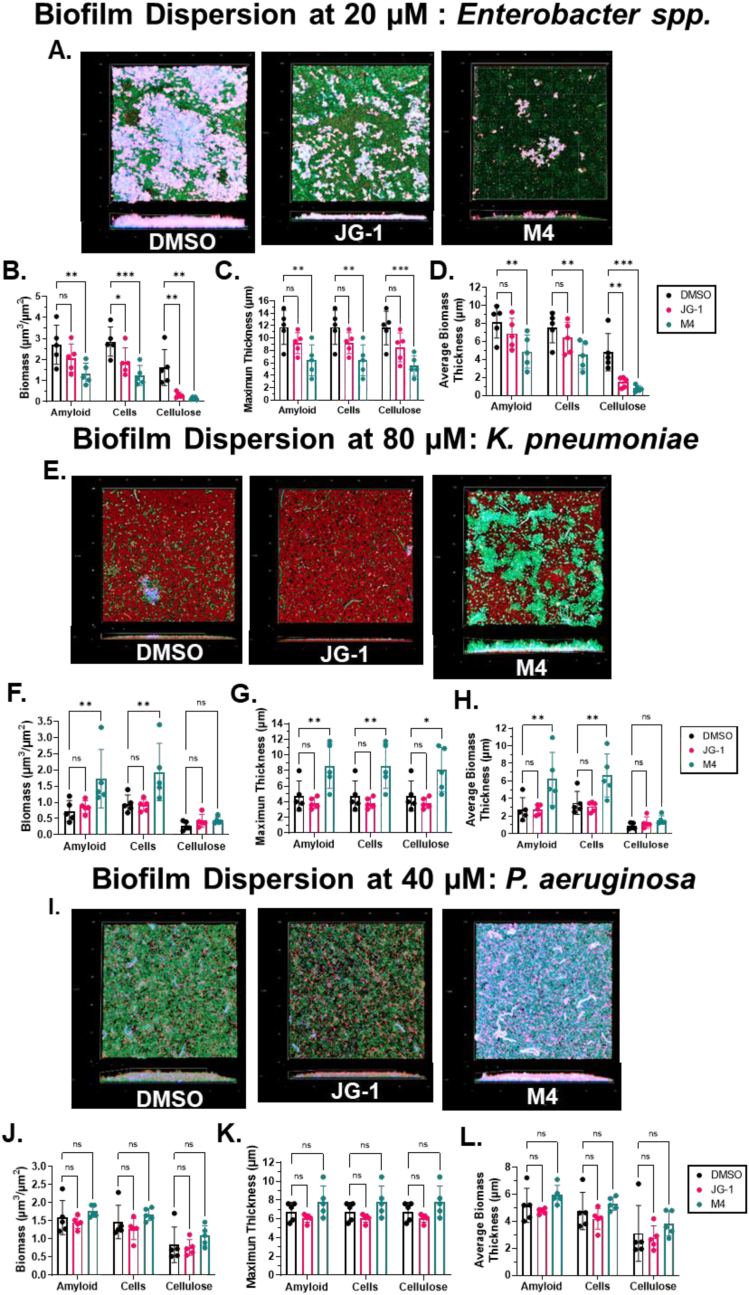
In order to detect changes in biofilm structure associated with dispersal activity of the lead compounds, pre-formed biofilms of Enterobacter spp., K. pneumoniae, and P. aeruginosa were incubated with JG-1, M4, or DMSO for 24 h prior to labeling for cellulose, amyloid, and bacterial cells (Fig. 5A, E, and I.). In Enterobacter spp., we observed a significant decrease in biomass, average thickness, and maximum thickness of all three biofilm components in the biofilms of cells treated with M4 (Fig. 5B-D.). We observed significant decreases of biofilm cells and average thickness of cellulose of cells treated with JG-1 and non-significant decreases of all other components (Fig. 5B-D.). In K. pneumoniae, we observed a significant increase in the biomass, average and maximum thickness of amyloid and biofilm cells, and the maximum thickness of cellulose of biofilms treated with M4 (Fig. 5F-H.). We observed no measurable difference between any other components of biofilms treated with JG-1. As expected, in P. aeruginosa biofilms treated with JG-1/M4, we did not observe any differences between biofilm components treated compared to vehicle (Fig. 5J-L.). Thus, these results were generally consistent with the observed biofilm dispersion activity of JG-1 and M4 on Enterobacter spp. and the lack of either M4 or JG-1 activity on P. aeruginosa or K. pneumoniae. Overall, the confocal microscopy dispersion and inhibition results validated those observed in the crystal violet assays.
Fig. 5.
Evaluation of changes in biofilm structure associated with JG-1/M4 dispersal activity. (A-D) Enterobacter spp., (E-H) K. pneumoniae and (I-L) P. aeruginosa biofilms were grown in 8-well chambered coverglasses for 24 h before being exposed to 20 μM, 80 μM, or 40 μM of compound or vehicle (DMSO), respectively, for an additional 24 h (A, E, and I) Biofilms were stained with SYTO9, calcofluor white, and a human α-amyloid antibody in order to visualize cells (green), cellulose (blue), and curli fimbriae/amyloid (red), respectively, prior to fixation in 4% PFA. Representative z-stacks (N =5 per treatment) were captured at 63X magnification using a Zeiss LSM 800 laser scanning confocal microscope. (B-D, F-H, and I-L) COMSTAT2 software was used to calculate the (B, F, and J) biomass, (C, G, and K) biofilm maximum thickness, and (D, H, and L) biomass average thickness of biofilm z-stacks for each individual component (cells, cellulose, and amyloid). Data are presented as the mean +/- SD. Annotations above the data denote values that are significantly different from the vehicle control (DMSO), as determined by two-way ANOVA with Dunnett’s test for multiple comparisons; * = p < 0.05, ** = p < 0.01, *** = p<0.001, ns= not significant (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2.4. Effect on planktonic cells
Finally, we conducted additional planktonic cell viability assays to establish if there was a decrease in the planktonic cell populations when each bacterial species was treated with either JG-1 or M4 in comparison to the vehicle control. These experiments will establish if the observed inhibitory or dispersion effects of the two compounds are due to stasis of bacterial metabolism or due to bacterial cell death. Planktonic cells of A. baumannii and P. aeruginosa were exposed to high (80 μM) and low (10 μM) concentrations of JG-1 or DMSO control for 0–6 h (Fig. 6A.) and MRSA and E. faecium to high (80 μM) and low (5 and 10 μM) concentrations of M4 or DMSO for 0–24 h (Fig. 6B.). After exposure, CFUs/mL were determined at various time points and compared to the DMSO treated cells. No difference was observed between A. baumannii or P. aeruginosa cells treated with DMSO or JG-1 at all time points. This suggests that when under non-biofilm inducing conditions, the compounds have no effect on planktonic cell metabolism or viability, but that under biofilm-inducing conditions, JG-1 has a bacteriostatic effect on planktonic cells, limiting their ability to produce biofilm.
Fig. 6.
Planktonic cell viability after treatment with lead compounds. (A) Evaluation of antibiotic activity of JG-1 against A. baumannii and P. aeruginosa. Liquid cultures of A. baumannii and P. aeruginosa were treated with low (10 μM) and high (80 μM) concentrations of M4 for 2–6 h before being transferred to LB agar without compound and incubated at 37 °C overnight. The colony forming units (CFU) remaining after exposure were counted and compared to vehicle. (B) Evaluation of bactericidal activity of M4 against S. aureus (MRSA) and E. faecium CI#1. Liquid cultures of MRSA and E. faecium were treated with low (5 μM and/or 10 μM) and high (80 μM) concentrations of M4 for 2–24 h before being transferred to LB agar without compound and incubated at 37 °C overnight. The CFUs remaining after exposure were counted and compared to vehicle. Bars represent average and standard error of the mean of all replicates. Annotations above the data (MRSA * = p < 0.05 and E. faecium # = p < 0.05) denote values that are significantly different from the vehicle control as determined by multiple t-tests and correction for multiple comparisons using the Holm-Šídák method. All other comaprisons are non-significant. N = 2–5 biological repeats each demonstrating similar relative differences between strains and conditions.
We observed a reduction in viable cells of MRSA and E. faecium after 4–6 h of treatment with the 10 μM dose of M4 in comparison to cells treated with DMSO. Total loss of viable cells was seen with MRSA at the 10 μM dose and both MRSA and E. faecium at the 80 μM dose of M4 at 24 h. No loss of viable cells was observed in cells treated with a 5 μM dose of M4 in MRSA or the 10 μM dose of M4 in E. faecium after 24 h in comparison to the vehicle. Our results demonstrate that the reduction in biofilm formation we observed previously is actually a result of a bactericidal effect of M4 on S. aureus and E. faecium and not specific to the biofilm phenotype.
4. Discussion
The great variability of conditions that promote optimal biofilm formation between each species in the ESKAPE group is not surprising given the genetic and environmental heterogeneity of these organisms. Similarly not surprising are the varied responses of different species to the same in vitro environment. For example, A. baumannii and Enterobacter spp. biofilm production was tested in the same conditions and yet produced very different results: A. baumannii biofilms averaged OD570 > 2.5 and Enterobacter spp. biofilms averaged much lower at OD570 < 1.1. Similarly to our results, other studies have found that even within the same species there were varied responses to the same environment [23]. Notably, the E. coli clinical isolates produced biofilms in TSB and LB but MC4100 and the enteroaggregative strain failed to produce biofilms in any of the conditions tested.
Biofilms are generally produced as a stress response to a hostile environment, often due to unfavorable cell density, nutrient levels, temperature, pH, or osmolality [3]. It is believed that pockets within the biofilm can act as storage for nutrients and that channels may facilitate dissemination of nutrients and oxygen more effectively than if the cells lived independently [24,25]. Accordingly, Enterobacter spp. produced the most biofilm in LB and TSB that was diluted 1:20 with dH2O and incubated at 30 °C. Low nutrients and a low temperature may be interpreted as a stressful environment, promoting the biofilm state over a planktonic phenotype. However, other species produced their largest biofilms in high-nutrient conditions which would typically be expected to be more favorable for a planktonic state. For example, S. aureus only produced significant amounts of biofilm when incubated in TSB supplemented with extra glucose. S. aureus can use glucose as an energy source, producing acidic byproducts and thereby acidifying the media [26]. S. aureus is known to activate the production of fibronectin-binding proteins (FnPs) in low-pH environments and FnPs promote aggregation of cells into biofilms [27,28]. TSB normally contains glucose (2.5 g/L) but the increased amount of glucose in the supplemented media may be causing enough acid-stress to trigger FnP-based biofilm formation in our assay.
While other studies suggested that Enterococcus biofilms can form in TSB supplemented with glucose [29], this media did not support significant biofilm formation in any of our isolates. Similarly we could not produce robust biofilms using M17 broth [30]. Only when TSB was supplemented with both glucose and hemin or M17 was supplemented with FeSO4 did we see a dramatic increase in biofilm production. Enterococcus is commonly isolated from the gastrointestinal tract where competition for iron between microbes and the host is fierce. Enterococci and many other bacteria found in the GI-tract can use iron for electron transfer and energy production [31]. Enterococci are unique in that they are very iron tolerant and can withstand iron concentrations that are toxic to many other species, providing a competitive advantage [25]. It was previously described that Enterococci sequester iron in pockets in their biofilm, specifically generating more biofilm mass in TSBG supplemented with of FeCl3, FeSO4, and Fe2(SO4)3 [25]. The GI-tract is also a low oxygen environment in which organisms compete for oxygen, which is necessary in more efficient aerobic respiration. Enterococci encode a cytochrome-bd oxidase that they use as a terminal electron acceptor, with heme as its cofactor, during aerobic respiration [32]. However, Enterococci do not produce porphyrin and therefore cannot produce heme, necessitating a reliance on exogenous heme in order to use this pathway [25]. By supplementing TSBG and M17 with hemin or FeSO4, respectively, we enable our E. faecium isolates to benefit from this energy-saving pathway which appears to aid in biofilm production.
Cell density is also known to influence biofilm formation in organisims such as Vibrio cholerae [33] and S. aureus [34], which are known to produce less biofilm when they perceive a high cell density in the environment. Notably in Enterobacter spp. and S. aureus conditions that promoted more biofilm formation tended to have fewer planktonic cells present (Supplementary Figs. 1A and B.). The opposite was true for A. baumannii and E. faecium, in which increased numbers of planktonic cells corresponded to higher detected biofilm mass (Supplementary Figs. 1D and G.). In K. pneumoniae, P. aeruginosa, and E. coli, cell density did not seem to be associated with biofilm mass (Supplementary Figs. 1C, E, F.). Our findings are in contrast to reports that E. coli and P. aeruginosa increased biofilm mass is initiated by high cell density [35]. However, these differences may be explained in part by differences in testing conditions. Thus, the ESKAPE pathogens may respond differently to other stressors, such as cell density when grown in optimal in vitro biofilm development conditions.
To maximize the utility of our assays, we also wanted to identify isolates within a species that produced more biofilm than their counterparts. It was determined that K. pneumoniae ATCC 13883, Top52, and IA565 produced different amounts of biofilm when grown in the same conditions. Klebsiella biofilms contain a large amount of polysaccharides and have a slimy consistency [36,37], which may be detrimental for use in our rapid attachment assay design. This slime may result in a weaker attachment of the biofilm to the 96-well plates, allowing them to be more easily washed away. Consistent with this, of the K. pneumoniae isolates we tested, colonies of IA565 are the least mucoid on agar, likely representing a lower polysaccharide content, and were the best biofilm formers. Other ESKAPE species’ biofilm formation may also depend on exopolysaccharide production, or may involve different biofilm- or metabolic-associated factors.
The problem of chronic biofilm-mediated infections has persisted due to a lack of available drugs or techniques that can disperse or kill bacteria within biofilms. Anti-biofilm drugs may be part of the arsenal necessary to address this problem; however, there is little consensus on the method for testing these drugs in vitro. To address this problem, we have developed model assays that can be used to test anti-biofilm compounds against members of the ESKAPE group, a major cause of antibiotic resistance and chronic biofilm-mediated infections in the clinic. By growing biofilms with appreciable biomass in 96-well plates, users can quickly screen many compounds and analyze both small and large differences in biofilm mass that can differentiate a successful candidate from an unsuccessful one.
In initial Salmonella studies, we screened small molecule libraries for promising hits to test against Salmonella biofilms using our in vitro biofilm assay [19]. We identified two small molecules, JG-1 and M4, that inhibit and disperse Salmonella biofilms in vitro and in vivo [15]. While activity in Salmonella is significant on its own, we wanted to determine their breadth of activity and potential therapeutic uses in other pathogenic biofilm-associated bacterial species. Thus, we employed our ESKAPE biofilm assays in order to determine the in vitro anti-biofilm effects of JG-1 and M4. We identified that our lead compounds have significant anti-biofilm activity against Enterobacter spp., P. aeruginosa, and A. baumannii. JG-1 inhibits biofilm formation in three out of six species tested (Enterobacter spp., A. baumannii, and P. aeruginosa) and M4 inhibits biofilm formation in all of the species tested with the exception of K. pneumoniae. In addition to their inhibitory effects, JG-1 and M4 also disperse Enterobacter spp. biofilms, and JG-1 disperses P. aeruginosa biofilms at higher concentrations (>10 μM). Importantly, the lack of an inhibitory effect on planktonic cells in these species suggests that the compounds antagonize biofilm formation without antagonizing growth of planktonic cells. This lack of growth restriction may limit selective pressure to develop resistance which is commonly seen in antibiotics and limits their long term use. In contrast, we observed that M4 had a bactericidal effect on S. aureus and E. faecium. These results were somewhat suprising given the lack of growth attenuation in other species. However, we believe that the antibiotic effects of M4 represent a new avenue for use in treating multi-drug resistant S. aureus and E. faecium infections. Introducing a drug with both antibiofilm and antibiotic effects is important given the increasingly limitied treatment options for patients due to the prevalence of antibiotic resitance in S. aureus and E. faecium. In this way the antibiotic activity of M4 further expands the utility and impact of this compound. These are promising data that suggest that our compounds may be utilized to treat chronic infections caused by pathogens in addition to Salmonella.
In our crystal violet dispersion assays, we noted some increase in biofilm mass in response to treatment of K. pneumoniae, S. aureus, and P. aeruginosa biofilms with M4. We do not know why this occurs but we speculate that it may be related to stress response. It is known that exposing biofilms to subinhibitory concentrations of antibiotics leads to increased biofilm formation [38]. The mechanisms by how this occurs vary, but it is generally believed to be a stress response to environmental cues [[38], [39], [40], [41]].
While the crystal violet-based assays we used to evaluate compound efficacy in our screen allow for efficient and accurate measurement of changes in biofilm mass, we wanted to also identify changes in biofilm structure as a result of JG-1/M4 treatment. We previously examined JG-1 and M4's effects on cellulose, curli (amyloid), and cells and noted that after compound treatment, Salmonella biofilms showed a decrease in biomass and thickness in cells, curli, and cellulose [15]. Given the diversity in extracellular matrix components between species and our previous study, we aimed to detect cellulose, amyloid, and cells, as these are known to be important for biofim formation in the ESKAPE pathogens as well [[42], [43], [44], [45], [46], [47]]. Curli fimbriae are composed of amyloid proteins and are a major component of Salmonella, E. coli, P. aeruginosa, and Enterobacter spp. biofilms [48,49]. In addition to curli fimbriae, P. aeruginosa also produce other amyloid proteins involved in aggregation and biofilm formation [42]. While S. aureus, K. pneumoniae, and E. faecium do not produce curli, biofilms of these species are known to contain functional amyloid proteins [37,42] In the confocal microscopy assays, we observed a general agreement in inhibition and disruption of biofilms compared to the crystal violet assays. As expected, in Enterobacter spp. we observed the same trends in change in biofilm components in both inhibition and dispersion assays, notably a decrease in amyloid, cells, and cellulose. We also noted that when K. pneumoniae biofilms and P. aeruginosa cells were treated with M4, we observed an increase in the average biomass thickness of amyloid. Considering the dramatic difference in abundance and thickness of each biofilm component between species, it is interesting that the these similar trends were observed.
Biofilm inhibition by JG-1 and M4 in some species but not others suggests that the target of action of each compound is present in some but not all isolates tested. Additionally, it is unclear if the same target (with the same function) is present in each affected organism, but we can infer that the potential target affects biofilm formation in some microbes but is lethal in others. The effects of our compounds on the tested ESKAPE group pathogens are summarized in Supplementary Table 2. Further mechanism of action studies are needed to better understand the anti-biofilm function of our lead compounds in Salmonella, Enterobacter spp., S. aureus, E. faecium, P. aeruginosa, and A. baumannii, as well as their anti-bacterial effect in S. aureus and E. faecium. This will enable us to further modify these compounds to optimize their activity.
Funding
This work was supported by the National Institutes of Health (R01 AI116917; R01 AI116917-07A1S1), the NCH Infectious Diseases Consortium, and an Ohio State Infectious Diseases Institute Transformative Trainee award.
CRediT authorship contribution statement
Aliyah N. Bennett: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Roles/. Katherine J. Woolard: Conceptualization, Data curation, Investigation, Writing – review & editing. Amy Sorge: Conceptualization, Data curation, Investigation, Writing – review & editing. Christian Melander: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. John S. Gunn: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Roles/.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Christian Melander reports a relationship with Agile Sciences Inc that includes: board membership. John Gunn has patent #US20230138583A1 pending to North Carolina State, University Ohio State Innovation Foundation.
Acknowledgements
We thank Lauren Bakaletz for providing us with many of the isolates we used in this study. We thank Jeremiah Johnson for providing us with the K. pneumoniae Top52 and IA565 isolates and his guidance in using them. We also would like to thank the Ohio State University Hospital East Clinical Microbiology Lab for providing us with the utilized clinical isolates, and Erin M. Vasicek for help with confocal microscopy.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2023.100158.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Guilhen C., Forestier C., Balestrino D. Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol Microbiol. 2017;105:188–210. doi: 10.1111/mmi.13698. [DOI] [PubMed] [Google Scholar]
- 3.Fux C.A., Costerton J.W., Stewart P.S., Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Sharma D., Misba L., Khan A.U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8:76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gene expression in Pseudomonas aeruginosa biofilms | Nature. https://www.nature.com/articles/35101627 [DOI] [PubMed]
- 6.Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis | applied and environmental Microbiology. https://journals.asm.org/doi/full/10.1128/AEM.67.7.2958-2965.2001 [DOI] [PMC free article] [PubMed]
- 7.Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli | journal of bacteriology. https://journals.asm.org/doi/full/10.1128/JB.181.19.5993-6002.1999 [DOI] [PMC free article] [PubMed]
- 8.Tomaras A.P., Dorsey C.W., Edelmann R.E., Actis L.A. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology (Read) 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 9.Chua S.L., et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun. 2014;5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 10.Darouiche R.O., Mansouri M.D., Gawande P.V., Madhyastha S. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J Antimicrob Chemother. 2009;64:88–93. doi: 10.1093/jac/dkp158. [DOI] [PubMed] [Google Scholar]
- 11.Hubeny J., Korzeniewska E., Ciesielski S., Płaza G., Harnisz M. The resistome of ESKAPEE pathogens in untreated and treated wastewater: a polish case study. Biomolecules. 2022;12:1160. doi: 10.3390/biom12081160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savin M., et al. ESKAPE bacteria and extended-spectrum-β-lactamase-producing Escherichia coli isolated from wastewater and process water from German poultry slaughterhouses. Appl Environ Microbiol. 2020;86 doi: 10.1128/AEM.02748-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice L.B. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol. 2010;31:S7–S10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 14.Donlan R.M. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandala J.L., et al. A dual-therapy approach for the treatment of biofilm-mediated Salmonella gallbladder carriage. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pletzer D., Mansour S.C., Hancock R.E.W. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia J., et al. Antimicrobial, antibiofilm, and antioxidant properties of boletus edulis and neoboletus luridiformis against multidrug-resistant ESKAPE pathogens. Front Nutr. 2022;8 doi: 10.3389/fnut.2021.773346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulani M.S., Kamble E.E., Kumkar S.N., Tawre M.S., Pardesi K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woolard K.J., Sandala J.L., Melander R.J., Gunn J.S., Melander C. Development of small molecules that work cooperatively with ciprofloxacin to clear salmonella biofilms in a chronic gallbladder carriage model. Eur J Med Chem. 2022;232 doi: 10.1016/j.ejmech.2022.114203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vorregaard, M. Comstat2 - a modern 3D image analysis environment for biofilms..
- 21.O'Toole G.A. Microtiter dish biofilm formation assay. J Vis Exp. 2011 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandala J., Gunn J.S. In: Salmonella: methods and protocols. Schatten H., editor. Springer US; 2021. In vitro evaluation of anti-biofilm agents against Salmonella enterica. (127–139). [DOI] [PubMed] [Google Scholar]
- 23.Dadawala A.I. Assessment of Escherichia coli isolates for in vitro biofilm production. Vet World. 2010;3 [Google Scholar]
- 24.The Matrix Reloaded How sensing the extracellular matrix synchronizes bacterial communities | journal of bacteriology. https://journals.asm.org/doi/10.1128/JB.02516-14 [DOI] [PMC free article] [PubMed]
- 25.Keogh, D. et al. Extracellular electron transfer powers Enterococcus faecalis biofilm metabolism. mBio 9, e00626-17.. [DOI] [PMC free article] [PubMed]
- 26.Jurtshuk P. In: Medical Microbiology. Baron S., editor. University of Texas Medical Branch at Galveston; 1996. Bacterial metabolism. [PubMed] [Google Scholar]
- 27.McCarthy H., et al. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol. 2015;5 doi: 10.3389/fcimb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergara-Irigaray M., et al. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun. 2009;77:3978–3991. doi: 10.1128/IAI.00616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heikens E., Bonten M.J.M., Willems R.J.L. Enterococcal surface protein esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007;189:8233–8240. doi: 10.1128/JB.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristich C.J., Li Y.-H., Cvitkovitch D.G., Dunny G.M. Esp-independent biofilm formation by Enterococcus faecalis. J Bacteriol. 2004;186:154–163. doi: 10.1128/JB.186.1.154-163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh P.K., Parsek M.R., Greenberg E.P., Welsh M.J. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 32.Winstedt L., Frankenberg L., Hederstedt L., von Wachenfeldt C. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J Bacteriol. 2000;182:3863–3866. doi: 10.1128/jb.182.13.3863-3866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammer B.K., Bassler B.L. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 34.Vuong C., Saenz H.L., Götz F., Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 35.Passos da Silva D., Schofield M.C., Parsek M.R., Tseng B.S. An update on the sociomicrobiology of quorum sensing in gram-negative biofilm development. Pathogens. 2017;6:51. doi: 10.3390/pathogens6040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifi K., et al. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur J Microbiol. 2016;9 doi: 10.5812/jjm.30682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nirwati H., et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13:20. doi: 10.1186/s12919-019-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranieri M.R., Whitchurch C.B., Burrows L.L. Mechanisms of biofilm stimulation by subinhibitory concentrations of antimicrobials. Curr Opin Microbiol. 2018;45:164–169. doi: 10.1016/j.mib.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Turnbull L., et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun. 2016;7 doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaplan J.B. Antibiotic-induced biofilm formation. Int J Artif Organs. 2011;34:737–751. doi: 10.5301/ijao.5000027. [DOI] [PubMed] [Google Scholar]
- 41.Ranieri M.R.M., et al. Thiostrepton hijacks pyoverdine receptors to inhibit growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.00472-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen L.F.B., Schafer N., Wolf-Perez A., Madsen D.J., Otzen D.E. In: Perrett S., Buell A.K., Knowles T.P.J., editors. vols. 113–159. Springer; 2019. Bacterial amyloids: biogenesis and biomaterials. (Biological and bio-inspired nanomaterials: properties and assembly mechanisms). [DOI] [Google Scholar]
- 43.Matilla-Cuenca L., Toledo-Arana A., Valle J. Anti-biofilm molecules targeting functional amyloids. Antibiotics. 2021;10:795. doi: 10.3390/antibiotics10070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller A.L., Bessho S., Grando K., Tükel Ç. Microbiome or infections: amyloid-containing biofilms as a trigger for complex human diseases. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.638867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C., Lau C.Y., Ma F., Zheng C. Genome-wide screen identifies curli amyloid fibril as a bacterial component promoting host neurodegeneration. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2106504118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kosolapova A.O., Antonets K.S., Belousov M.V., Nizhnikov A.A. Biological functions of prokaryotic amyloids in interspecies interactions: facts and assumptions. Int J Mol Sci. 2020;21:7240. doi: 10.3390/ijms21197240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., et al. Increased biofilm formation ability in Klebsiella pneumoniae after short-term exposure to a simulated microgravity environment. Microbiol. 2016;5:793–801. doi: 10.1002/mbo3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tursi S.A., Tükel Ç. Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, and disease implications. Microbiol Mol Biol Rev. 2018;82 doi: 10.1128/MMBR.00028-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zogaj X., Bokranz W., Nimtz M., Römling U. Production of cellulose and curli fimbriae by members of the family enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.