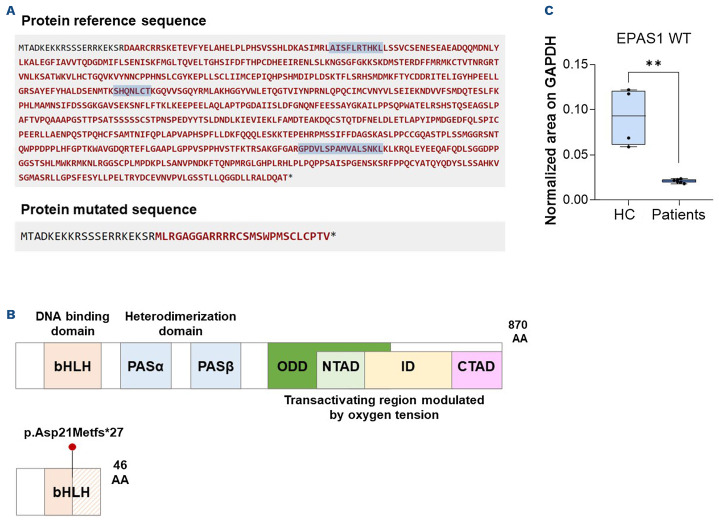
Figure 2.
The EPAS1 deletion variant accounts for EPAS1 haploinsufficiency. (A) Predicted protein reference (upper) and mutated (lower) sequence by Mutalyzer (v 3.0.4, https://mutalyzer.nl/). The three EPAS1 wildtype-derived peptides for multiple reaction monitoring analysis are highlighted in light blue. (B) General protein model of EPAS1 representing EPAS1 wildtype (upper) and hypothesized mutated (lower) protein structure. (C) Boxplot showing normalized mean areas for the amount of EPAS1 wildtype protein in patients compared to that in healthy controls. The mean of the total transition area relative to each peptide of the EPAS1 protein was normalized to the mean of the total transition area of the GAPDH protein. All samples were run in technical duplicates. **P<0.01. P value by the Mann-Whitney test.