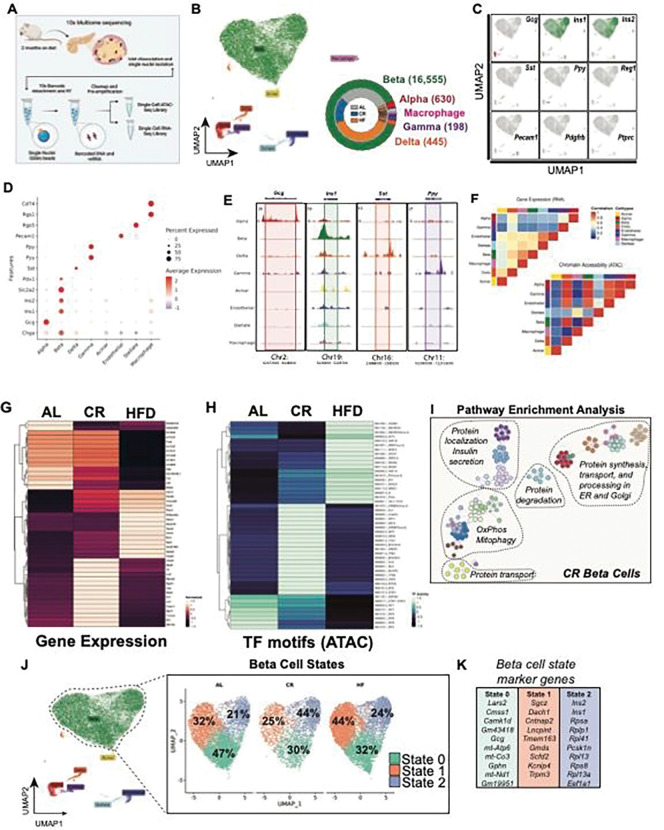
Figure 2. Single cell multiome sequencing of islets reveals diet-specific changes to beta-cell heterogeneity.
(A) Schematic representation of the workflow used for single nuclei multiome sequencing (ATAC + RNA) of pancreatic islets isolated from AL, CR and HFD mice after 2 months on diet. (B) Uniform Manifold Approximation and Projection (UMAP) of the integrated transcriptome and chromatin dataset. Data from a total of n=18,741 islet cells from n=2 mice per diet group. Different cell types are indicated by various colors and labels. Inset, donut plot with the total number of cells in each cell cluster. (C) UMAPs showing the expression of cell marker genes. (D) Dot plot with relative expression of marker genes in alpha, beta, delta, gamma, acinar, endothelial, stellate and macrophage cell clusters. (E) Genome tracks showing ATAC peaks for hormone markers: Gcg (alpha cell), Ins1 (beta cell), Sst (delta cell) and Ppy (gamma cell). (F) Heatmap showing the correlation between gene expression (RNA-seq) and chromatin accessibility (ATAC-seq) across the identified cell types. (G) Heatmap with the top differentially expressed genes (RNA-seq) and (H) top ATAC-seq peaks in transcriptional factors (TF) motifs in beta-cells from AL, CR, and HFD mice. (I) Annotated node map with pathway enrichment analysis of differentially expressed genes and TF motifs in beta-cells from CR mice. Analysis was performed using Metascape with an FDR < 0.05. (J) UMAP projection of beta-cells and identified subclusters from AL, CR and HFD mice. Beta-cell populations were defined according to distinct transcriptional states. (K) Top marker genes from each beta-cell state.