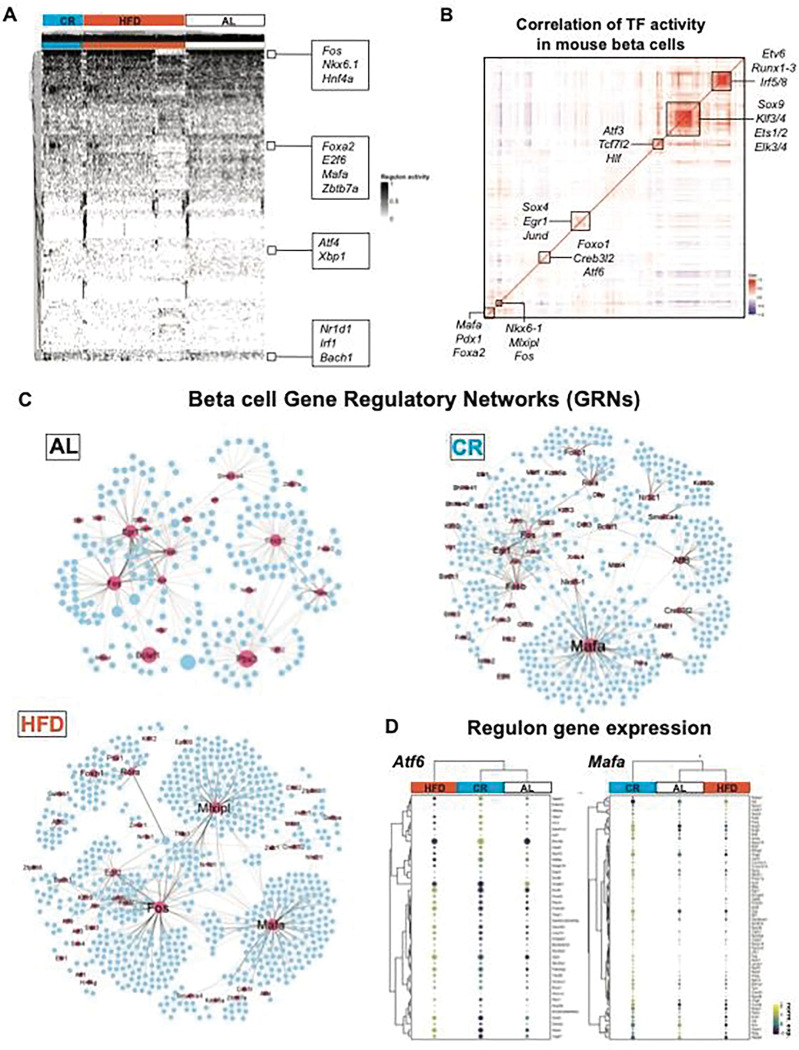
Figure 3. CR reprograms beta-cell gene regulatory networks (GRNs).
(A) SCENIC heatmap with hierarchical clustering analysis of TF activity in beta-cells from AL, CR and HFD mice after 2 months on diet. TFs identified as “ON” are shown in black, while TFs identified as “OFF” are in white. (B) Pearson correlation matrix of n=272 TF identified in all mouse beta cells (all diet groups together). Boxes highlight clusters of TFs with high degree of correlation. (C) Gene regulatory networks (GRNs) formed by TFs identified using SCENIC in beta-cells from AL, CR and HFD mice. TFs are shown as pink nodes, while target protein-coding genes are shown in blue. Node size represents the “betweenness centrality” measurements that report in the influence of a given TF within a network. (D) Dot plot and hierarchical clustering showing the gene expression levels of target genes associated to Atf6 and Mafa GRNs from AL, CR and HFD beta cells. Dot plot scale shows the relative expression level and percentage of cells expressing a target gene.