Figure 4: Transcription factors controlling differential gene expression in MG and MGPC following LD and NMDA treatment.
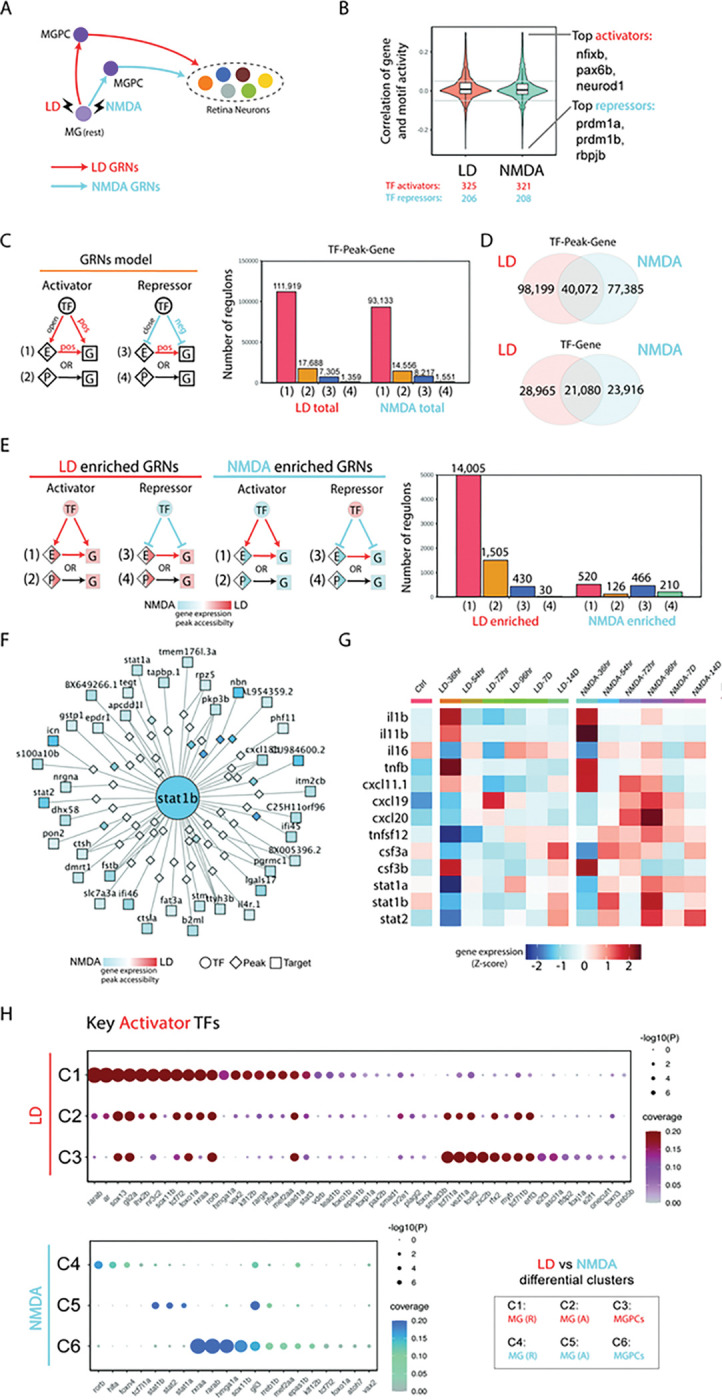
(A) Schematic of MG regeneration after LD and NMDA treatment.
(B) Inference of activator and repressor function for each individual transcription factor from mulitomic datasets. The y-axis represents the correlation distribution between gene expression and chromVAR score. The top three activator and repressor TFs are shown on the right.
(C) Gene regulatory networks of LD and NMDA datasets. (left) Triple regulons model. (right) barplot shows the types of regulons between LD and NMDA datasets.
(D) Venn diagram shows the overlap of regulons between LD and NMDA datasets.
(E) Enriched gene regulatory networks of LD and NMDA treatment. (left) Enriched Triple regulons model for each condition. (right) barplot shows the number of different types of regulons between LD and NMDA enriched GRNs.
(F) An example of stat2 regulons. Color indicates the log2 fold change between the LD and NMDA datasets.
(G) Heatmap showing the differentially expressed genes in microglia/macrophages after LD and NMDA treatment. Microglia/macrophage cells are ordered by time points after injury, and an averaged expression level is shown for each timepoint. Color represents mean-centered normalized expression levels.
(H) Dotplot showing the key activator TFs for each divergent gene cluster. The size of the dot shows the gene expression ratio, and color indicates the statistical significance of differential expression.
