Figure 5. Neurons and endothelial cells modify the splicing of mRNAs in astrocytes.
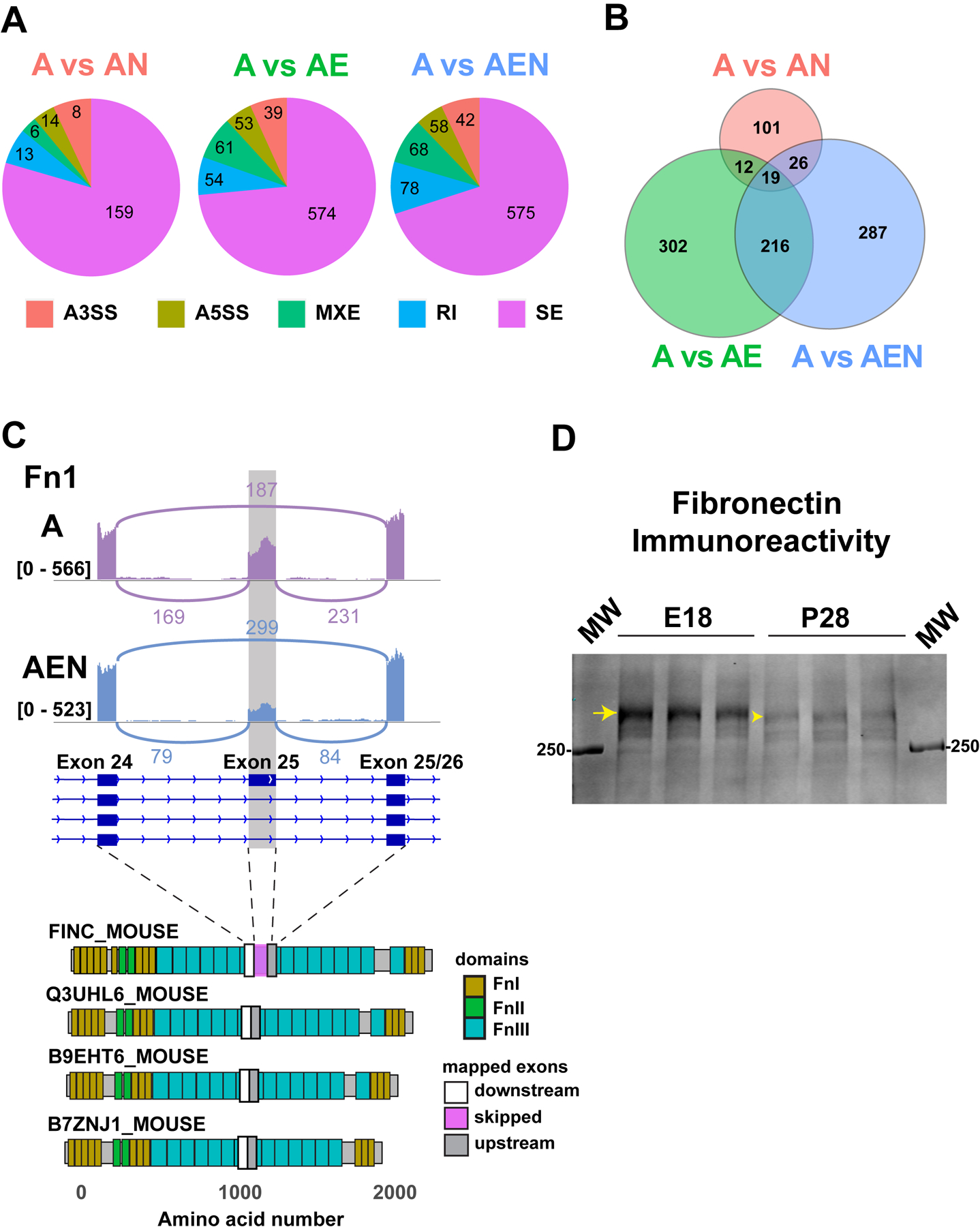
A. Frequencies of the five types of alternative splicing differentially detected in astrocytes monocultures versus astrocytes culture with neurons (A vs AN), endothelial cells (A vs AE), or with neurons and endothelial cells (A vs AEN). Five different types of alternative splicing were identified including alternative 3’ splice site (A3SS), alternative 5’ splice site (A5SS), mutually exclusive exons (MXEs), retained introns (RI), and skipped exons (SE). The numbers indicate the number of genes with differentially alternative events in the culture configurations indicated. Skipped exons were the most abundant alternative event. B. Scale vent diagram showing the unique versus shared skipped exons events in the different culture configurations. Only 19 skipped exon events were present in all co-culture configurations but absent in astrocyte monocultures. C. Fibronectin 1 (Fn1) is an example of a gene differentially regulated by neurons and endothelial cells through alternative splicing, specifically by exon skipping. The purple and blue traces represent raw data of the number of reads mapped to the Fn1 gene in astrocytes grown as monocultures and of astrocytes grown in the presence of neurons and endothelial cells respectively. The height of the purple and blue columns represents the number of reads. The gray column highlights exon 25. The transcript model of the Fn1 gene in dark blue is from the genome browser Ensembl (ENSMUST00000055226), boxes represent exons. D. Western blot analysis of mouse embryonic (E18) and adult (P28) brain cortex (tissue from three different animals each). Fibronectin bands were detected. The arrow highlights Fn1 long isoform (containing exon 25), while the arrowhead highlights Fn1 short isoform (lacking exon 25).
