Figure 1: Conserved host shutoff and ribosome binding of coronaviral Nsp1.
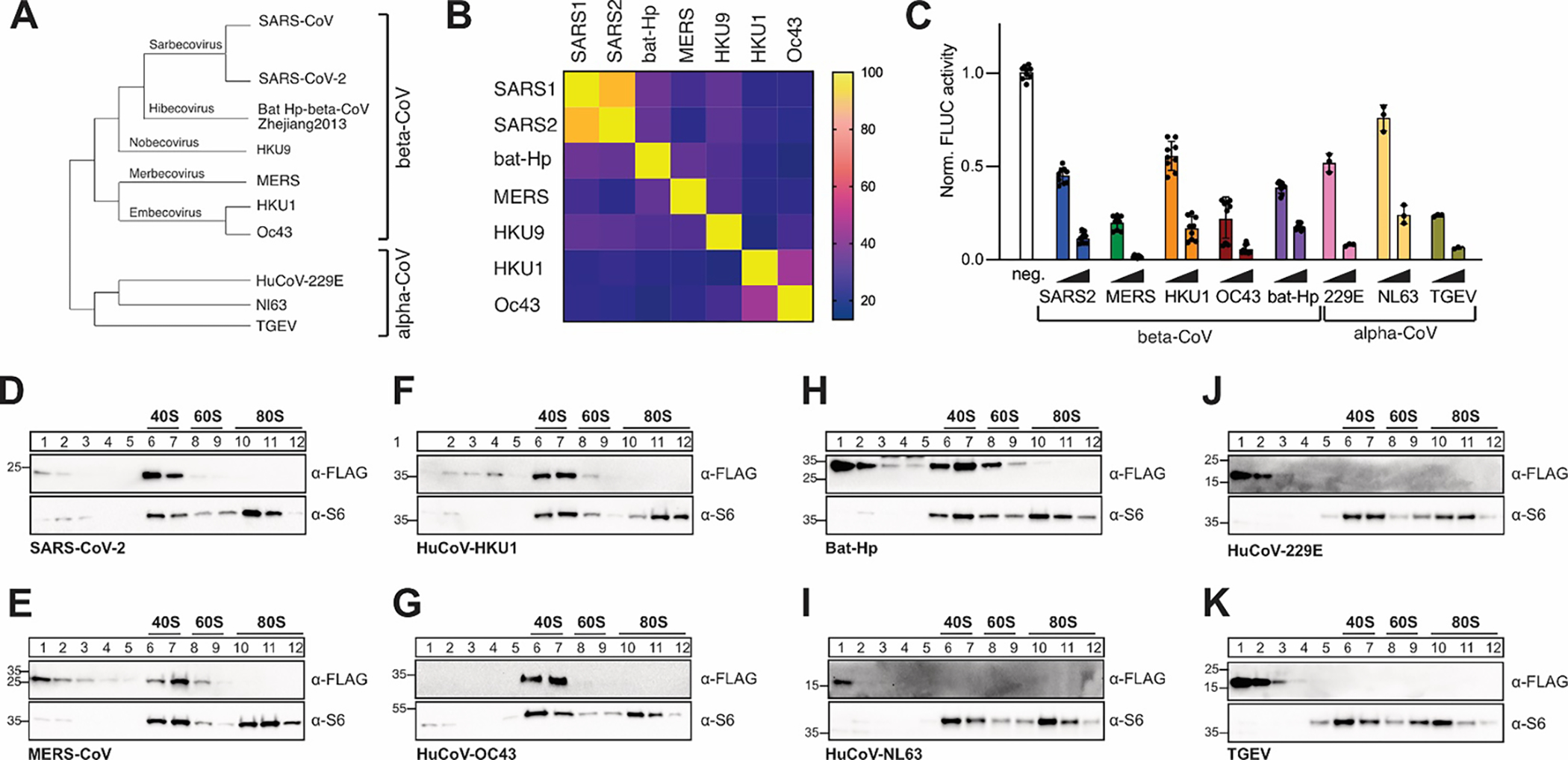
A) Evolutionary relationship of select α- and β-coronaviruses. The phylogenetic tree is based on RdRP conservation. B) Heat map of sequence variation in β-CoV Nsp1 proteins. C) Host shutoff in HEK293T cells transiently expressing Nsp1 from the indicated viruses, measured by luciferase activity of a co-expressed FLuc reporter gene. D-K) Sucrose gradient analysis of HEK293T cell lysates expressing 3x-FLAG tagged Nsp1 from SARS-CoV-2 (D), MERS-CoV (E), HuCoV-HKU1 (F), HuCoV-OC43 (G), Bat Hp β-CoV Zhejiang2013 (H), HuCoV-NL63 (I), HuCoV-229E (J) or TGEV (K). Western blot analysis for the presence of Nsp1 (α-FLAG) and ribosomal protein uS6 (α-S6).
