Figure 2: A conserved α-helical structure in the C-terminus of β-CoV Nsp1 mediates ribosome binding.
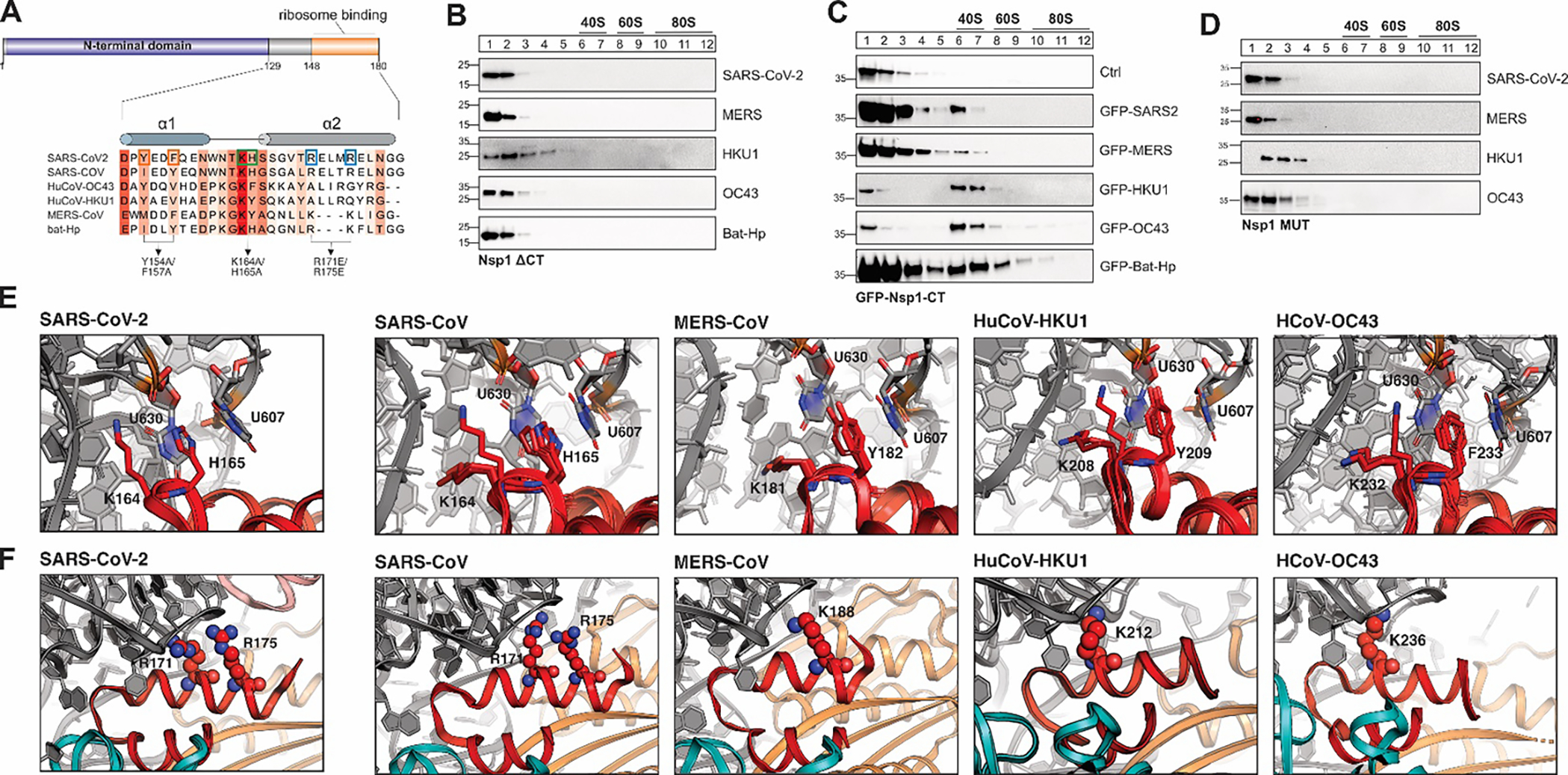
A) Sequence alignment of the C-terminal domain of Nsp1 from SARS-CoV, SARS-CoV-2, MERS-CoV, HuCoV-HKU1, HuCoV-OC43 and bat-Hp Nsp1. The Sequence conservation is indicated by red shading. Previously characterized mutants in the C-terminus of SARS-CoV-2 Nsp1 are indicated by boxes and arrows. B-D) Sucrose gradient analysis of HEK293T cells transiently expressing the indicated 3x-FLAG-tagged Nsp1 variants: C-terminally truncated Nsp1 mutants (B), GFP (top) or GFP fused to the CTD of Nsp1 from the indicated viruses (C), and Nsp1 variants in which the conserved KH or KY/F motif was mutated (D). Western blot analysis: α-FLAG. E) Details of the interactions between the KH-motif of the SARS-CoV-2 Nsp1 CTD and the ribosome (left, pdb 6zlw), and modeled interactions between the KH-motif of SARS-CoV (2nd panel), KY motif of MERS-CoV (3rd panel) and HuCoV-HKU1 (4th panel), and the KF-motif of HuCoV-OC43 Nsp1 (right) with the 18S rRNA. The aromatic residues of the KY/F-motif likely form stacking interactions with ribosomal U607 and U630, similar H165 in SARS-CoV-2 Nsp1. Nsp1 in red and 18S rRNA in grey. F) Details of the interactions between helix α2 of SARS-CoV-2 Nsp1 and the 18S rRNA of the small ribosomal subunit (left, pdb 6zlw). Details of the modeled interactions between helix α2 of Nsp1 from the indicated viruses and the 18S rRNA (right). Note that the electrostatic interactions of R171 and R175 are replaced by positively charged amino acids at different positions along α2 in MERS-CoV, HuCoV-HKU1 and HuCoV-OC43 Nsp1. Nsp1 is shown in red, 18S rRNA in grey and ribosomal proteins uS3 in teal and uS5 in orange.
