Figure 3: Correlation between ribosome binding and translation inhibition in different β-CoV Nsp1s.
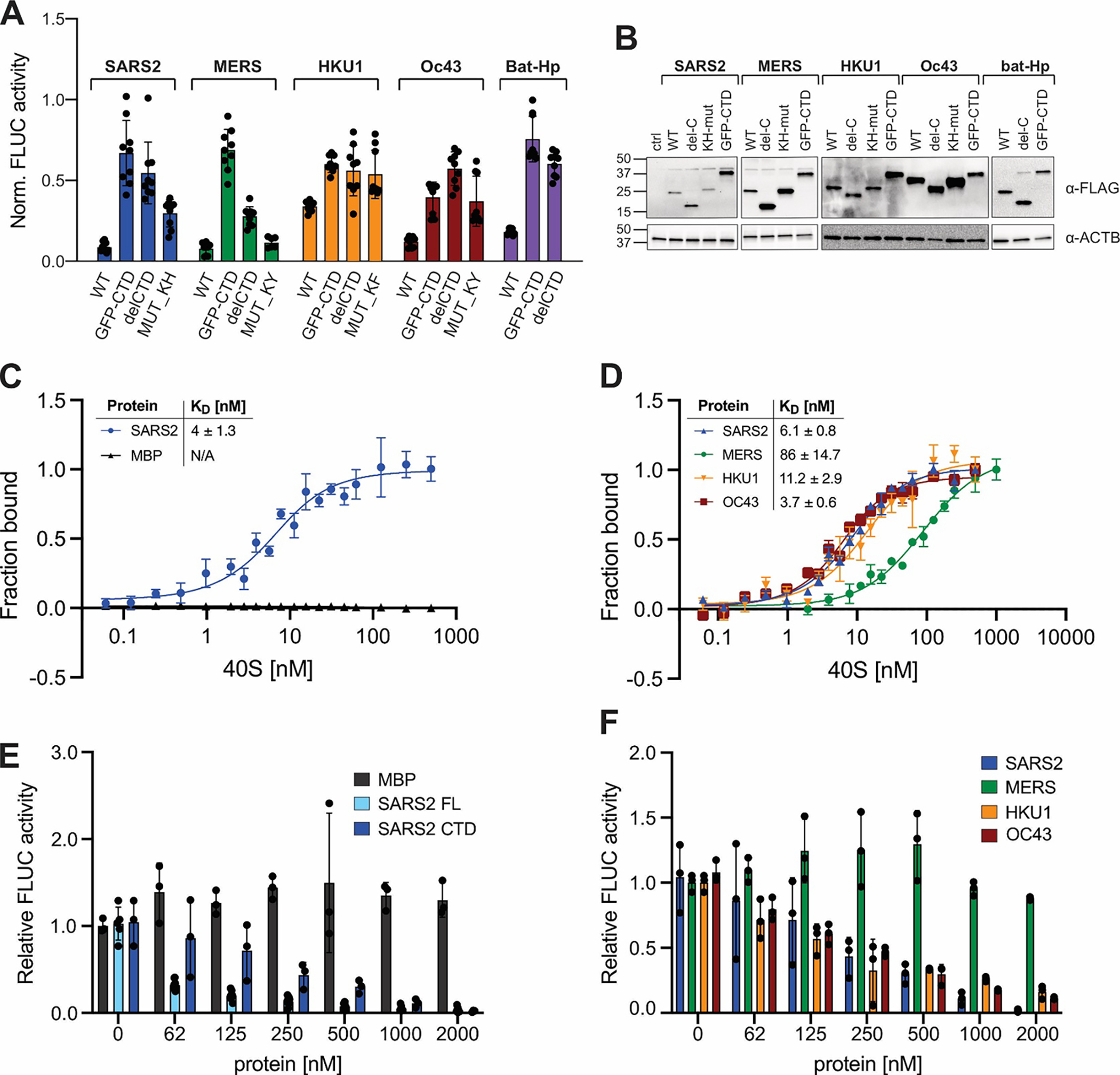
A) Host shutoff in HEK293T cells transiently expressing 3xFLAG-tagged wild-type or mutant Nsp1 from the indicated viruses, measured by FLuc activity. B) α-FLAG immunoblot analysis of HEK293T cells from (A). C) Equilibrium binding measurements of FAM-labeled SARS-CoV-2 Nsp1 to purified rabbit 40S ribosomal subunits, maltose binding protein (MBP) served as a negative control. n=3, error bars=SEM. D) Equilibrium binding measurements of FAM-labeled MBP fused to the Nsp1-CTD from the indicated viruses to rabbit 40S subunits. n=3, error bars=SEM. E) Cell-free translation of FLuc reporter mRNA in rabbit reticulocyte lysate supplemented with increasing concentrations of MBP, SARS-CoV-2 Nsp1 (“SARS2 FL”), or MBP fused to the CTD of SARS-CoV-2 (“SARS2 CTD”). n=3, error bars=SEM. F) Cell-free translation of an FLuc reporter mRNA in the presence of increasing concentrations of MBP fused to the Nsp1 CTD of the indicated viruses. n=3, error bars=SD.
