Abstract
The use of wastewater-based surveillance (WBS) for detecting pathogens within communities has been growing since the beginning of the COVID-19 pandemic with early efforts investigating severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) RNA in wastewater. Recent efforts have shed light on the utilization of WBS for alternative targets, such as fungal pathogens, like Candida auris, in efforts to expand the technology to assess non-viral targets. The objective of this study was to extend workflows developed for SARS-CoV-2 quantification to evaluate whether C. auris can be recovered from wastewater, inclusive of effluent from a wastewater treatment plant (WWTP) and from a hospital with known numbers of patients colonized with C. auris. Measurements of C. auris in wastewater focused on culture-based methods and quantitative PCR (qPCR). Results showed that C. auris can be cultured from wastewater and that levels detected by qPCR were higher in the hospital wastewater compared to the wastewater from the WWTP, suggesting either dilution or degradation of this pathogenic yeast at downstream collection points. The results from this study illustrate that WBS can extend beyond SARS-CoV-2 monitoring to evaluate additional non-viral pathogenic targets and demonstrates that C. auris isolated from wastewater is competent to replicate in vitro using fungal-specific culture media.
Keywords: Candida, C. auris, SARS-CoV-2, Wastewater, Wastewater-Based Surveillance, WBS
Graphical Abstract
1.0. Introduction
There is a paucity of fungal diseases among mammals compared to plants, amphibians, and insects.1 This resistance has been proposed to be the result of high temperatures which create a thermal restriction zone among humans. Most fungi do not grow well at high temperatures (>37°C) and prefer ambient conditions; however, Candida auris has been also observed to grow well at 42°C (108°F).2 Furthermore, C. auris’s capacity to survive in the clinical environment is a sign of how well adapted it is to living conditions outside of a host, including its resistance to various anti-fungal agents.3 For instance, C. auris has been recovered from salt marsh wetlands and sandy beaches of the Andaman Islands in the Indian Ocean,4, 5 and estuaries in northwest Colombia.6 In fact, C. auris can survive in environments several times saltier than the ocean.2
The capacity of C. auris for thermotolerance and halotolerance is ideal to persistently colonize patients’ skin, especially in areas such as the axilla and groin.2 This attribute coupled with the ability to survive on abiotic surfaces for prolonged periods7 has caused multiple nosocomial outbreaks in all the populated continents around the globe and caused the Centers for Disease Control and Prevention (CDC) to label it as an urgent threat.8,9 Moreover, transmission events were exacerbated by the COVID-19 pandemic as resources were diverted from C. auris surveillance to pandemic response. These changes in the US led to a 60% increase of C. auris clinical cases between 2019 to 2020 cases, according to the CDC.10 For example, C. auris outbreaks have been reported in COVID-19 units around the world,11, 12 including a Florida acute-care hospital that identified 35 patients (52%) to be colonized with C. auris. Furthermore, colonization can lead to invasive infections. For example, one study showed that out of 157 C. auris-colonized patients in an intensive care unit, 27 (17%) patients developed C. auris candidemia, and of these, seven developed a recurrent episode.13 Invasive infections can also be difficult to treat since C. auris is a multidrug-resistant organism which can be resistant to all classes of antifungals. Clusters of pan-resistance cases have been detected in Texas, District of Columbia, and New York.14 Risk factors for invasive infections and colonization include patients with indwelling medical devices, such as a tracheostomy and gastrotomy tubes, those who reside in ventilator-capable skilled nursing facilities and long-term acute care hospitals, and prior antimicrobial treatment.15
This increase in cases is not isolated in the US and is occurring globally. In the European Union, cases doubled between 2020 and 2021 and 11 new countries reported cases of C. auris.11 These events highlight how detection of C. auris is a necessary step to contain its rapid spread. Furthermore, data from the UK shows that early detection in combination with infection control and prevention measures, and screening can halt transmission of C. auris.11 Surveillance of C. auris is still a challenge in the United States.16 Some challenges include the lack of on-site testing and prolonged turnaround time when using public health laboratories. Consequently, C. auris has spread to 28 states in the US17 and it is likely existing in additional states that have not yet detected it. Furthermore, there is no global strategy for detection in developing nations as most laboratories use systems that misidentify C. auris such as Vitek 2 and API 20C.18 In 2018, nine years after C. auris was first detected, seven EU nations reported a lack of capacity to identify it.19 A follow-up survey in 2021 showed that four EU countries lacked national-level data for C. auris which raises the possibility of undetected transmission.11
The lack of global detection capacity can therefore lead to silent amplification of the organism and cause transmission, especially among patients in long-term acute care hospitals and ventilator-capable skilled nursing facilities. The paucity of detection capabilities do call for additional methods of surveillance.16, 18–20
Wastewater-based surveillance (WBS) can serve as a new powerful method of surveillance by enhancing C. auris detection for healthcare facilities by offering early warnings and real-time monitoring of transmission events, similarly to its use for detecting severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).21–24 It is recognized that C. auris is a very different microbe than SARS-CoV-2 (in terms of their phenotypic and morphological differences, different modes of infection, DNA detection as opposed to RNA detection, different modes of shedding, different shedding rates, and persistence in the environment). However, we do propose that WBS is a unifying approach as a means of detecting illnesses among a sewershed community, even for these different categories of microbes. WBS has unifying advantages in that it is not biased by individual human access to testing and willingness to report results.
WBS is also a cost-effective method to detect biomarkers (regardless of microbe type) during the early stages of colonization, even prior to symptoms of disease.25 Currently, the alternative is delayed by several days until a clinical culture is finalized. One study by Mataraci-Kara et al. (2020)26 examined the susceptibility patterns of various Candida species isolated from hospital wastewater and were able to determine that antifungal drug resistance did exist within wastewater and provided recommendations for future wastewater management.26 Comparatively, another study by Sabino et al. 201027 further confirmed the incidence of Candida species within an oncology hospital in Portugal illustrating clinical outcomes, antifungal susceptibility, and mortality occurrence in patients. To our knowledge, only two studies have recently been published bringing to light C. auris detection with WBS approaches performed by Barber et al. and Rossi et al..28, 29 Both studies, performed in Nevada, US, provided proof of concept that C. auris was detectable in wastewater with quantitative PCR (qPCR) assays, and that detections were higher in locations that service healthcare facilities following outbreaks in the hospital. Moreover, Rossi et al. further investigated the possibility of culturing C. auris from wastewater as there is reported evidence of culture-based recovery from marine waters.4, 29 The results from these prior studies provided grounds for additional experimentation within this study as C. auris detected in Nevada was measured from a different geographical and climatic setting than Miami, FL.
At the University of Miami (UM), the long-standing COVID-19 focused WBS program has been rapidly expanding to detect alternative targets within wastewater.30 C. auris has been of particular interest at the University hospital, as patients colonized by C. auris have represented a significant challenge for clinical management and safe discharge from the hospital. That is, it has been the case for the past 18–24 months that C. auris patients who had been admitted from skilled nursing facilities were successfully treated for the other maladies that led to their hospitalization, but then could not be discharged because the nursing facility would not accept C. auris patients. It was surmised that for an indeterminate number of these cases, the C. auris infection initially occurred at the external facility, i.e., before hospital admission. In either case, the presence of the multidrug-resistant fungus led to otherwise unnecessary and lengthy hospital stays.3
Understanding the levels of C. auris in the general community and within hospital settings can help healthcare providers to minimize spread. We hypothesize that prevalence of C. auris can be estimated from wastewater measurements, further investigating the results of previous studies.28, 29 We suspect that C. auris from infected or colonized patients within an acute care facility would ultimately be found in the sewage system, from either patient bathing or disposal of bodily waste and could be detectable with molecular assays and culturing. The intention of the current study is to build upon the efforts of Barber et al. and Rossi et al.. 28, 29 These prior studies focused on measurements at the wastewater treatment plant. We expand on these measurements by collecting wastewater samples from hospital specific sampling sites (manhole or clean out from the hospital building) and we also compared the wastewater results to available epidemiologic data at the hospital scale. This is the first time such comparisons are available due to the availability of hospital in-patient data.
2.0. Materials and Methods
2.1. Sample Collection and Study Design
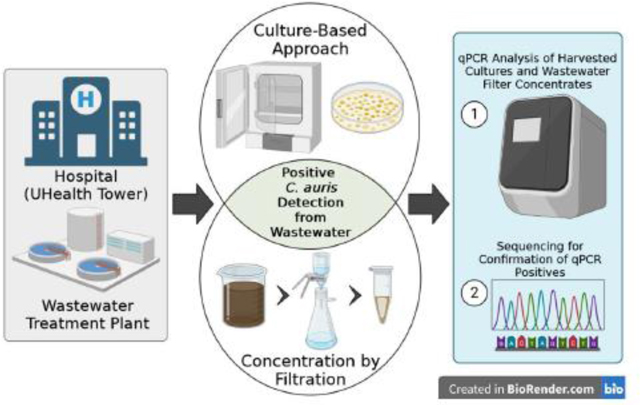
Wastewater samples, collected as grab samples from the University’s Hospital called UHealth Tower have been routinely collected weekly since September 2020 to measure the levels of SARS-CoV-2 RNA31 for the purpose of relating the number of clinical COVID-19 cases in the hospital against levels of RNA measured in wastewater. The two UHealth Tower hospital sampling sites included WM06 corresponding to a sewer lateral cleanout from the hospital building and another site, WM08, consisting of flow from a specific pipe within a manhole that transported only hospital wastewater. In addition, wastewater as 24-hour composites from the regional wastewater treatment plant (Central District Wastewater Treatment Plant (CDWWTP), sampling site WC0Dc, was also collected on the same weekly sampling schedule, as this treatment plant (located on the Virginia Key, FL) is the downstream processing plant for the University’s hospital. Two-liter samples were collected in the field. One liter of this sample was split and poured into a sterile HDPE bottle containing 0.1 g sodium thiosulfate (to reduce the chlorine residual), and immediately transported on ice packs to the Biospecimen Shared Resource Laboratory (BSSR) on the University’s Miller School of Medicine campus. Upon receipt at the laboratory, this 1-L sample was immediately split into two, 500 mL aliquots and set aside for the separate processing of C. auris and SARS-CoV-2.
Basic ambient (air temperature and humidity using NavClock app) and water quality parameters (pH, water temperature, salinity, specific conductivity turbidity, dissolved oxygen using a pre-calibrated Sonde, Xylem/YSI ProDSS) were measured in the field. These basic water quality parameters were measured in the second 1 L split of a 2 L sample retrieved from the sampling site. Summary statistics for these measurements are available in the supplemental text (Table S-1).
2.2. Concentration and Molecular Assessment for C. auris
The first 500 mL aliquot of wastewater analyzed for C. auris underwent sample processing optimized for DNA extraction. The sample was filtered with vacuum filtration (VF) using GN-6 Metricel membranes (Pall Corp. #66278, 47 mm diameter, 0.45 μm pore size) to clogging and the volume of the sample filtered was recorded. Following filtration, the filter was placed within a sterile petri dish and sliced in half with a disposable scalpel; one-half of the membrane was placed within a Zymo Research ZR BashingBead Lysis Tube (0.1 & 0.5 mm) (Cat#S6012–50) and topped with 1 mL of 1X DNA/RNA shield for immediate assessment, the other placed within a 5 mL conical tube in 1 mL of 1X DNA/RNA shield and stored at −80 °C.30 The filter was split in half due to the physical space limitations of the lysis tube and C. auris computations took into account filter splitting. All wastewater concentrate samples were kept at 4 °C until transported to the Center for AIDS Research (CFAR) for downstream nucleic acid extraction and follow up molecular analysis.
Nucleic acid extraction for C. auris included a bead-bashing step using an OMNI Bead-Ruptor 12 instrument. The half GN-6 filter membrane placed within the Zymo ZR BashingBead Lysis Tube was processed using the following bead-bashing protocol: 1 min. bashing, 5 min rest (x3 cycles, 18 minutes total). The initial centrifugation step (10,000 x g) following bead-bashing was increased by 2,000 x g from the manufacturer’s recommended protocol to improve the pelleting of ruptured membrane particles. A volume of 400 μL was used to purify DNA from samples according to the manufacturer’s protocol (ZymoBIOMICS DNA Miniprep Kit, Cat. # D4300). DNA in eluates were enumerated by qPCR assay which utilized previously published primers and reporter probe32 and a TaqMan Fast Universal PCR Master Mix (ThermoFisher Sci. Cat# 4352042) for quantification of the C. auris signal from wastewater. For validating the qPCR assays specificity for C. auris, a positive control was provided by the UM Hospital Pathology laboratory.
Additionally, two isolates derived from wastewater samples that tested qPCR-positive for C. auris were further evaluated by DNA sequencing. An amplicon larger than the qPCR amplicon was generated for sequencing and the resulting sequences were aligned to a partial sequence of ITS1 and the 28S rRNA gene and complete sequence of the 5.8S rRNA gene and ITS2 previously submitted to the National Center for Biotechnology Information (NCBI).
2.3. Concentration and Molecular Assessment for SARS-CoV-2
The second 500 mL aliquot of wastewater analyzed for SARS-CoV-2 was spiked with beta coronavirus-OC43 (500,000 viral particles per 500 mL) as the corresponding viral recovery control. Prior to utilizing OC43 as a control for measuring recovery efficiencies, several wastewater samples collected at different sites at different timepoints were evaluated for the presence of OC43 virion RNA. All of the samples were below detection limits for OC43 RNA by qPCR, providing confidence that the wastewater samples contain negligible amounts of background OC43 particles. From the analysis of OC43, recoveries were consistently near 22%. Results presented were not adjusted for recovery.
Upon spiking with OC43, the samples were subjected to electronegative (EN) filtration following the addition of magnesium chloride to a concentration of 50 mM. Hydrochloric acid (10%, Spectrum Chemical MFG Corp) was also added to this aliquot until the pH of the sample reached between 3.5 and 4.5. The purpose of these treatment reagents was to improve the binding affinity of viral particles to the negatively charged EN membranes (Millipore HAWP4700, 47 mm diameter, 0.45 μm pore size) by altering the chemical charge of ambient viral particles to positive allowing for binding of the viruses to the filter by charge, in addition to the capture of viruses associated with suspended particles larger than the filter pore size. Upon acidification of the wastewater, samples were filtered through the EN membranes to clogging and the volume was recorded. The EN membranes were folded in on themselves four times and immediately placed within 1.5 mL of 1X DNA/RNA shield (Zymo Research)24, 31, 33. For consistency, generated wastewater concentrate samples were also kept at 4°C until transport to the Center for AIDS Research (CFAR) for downstream nucleic acid extraction and follow up molecular analysis.
The RNA isolation process of wastewater concentrates at the CFAR laboratory utilized a modified protocol34 and the commercially available Quick-RNA™ Viral Kit (Zymo Research). The RNA extraction process of wastewater concentrates was optimized in 2020 at UM and performed for this study. In brief, the modifications of the manufacturer’s protocol included increases from 500 to 660 microliters of wash buffer volumes and 100% ethanol during the RNA extraction process, and a reduction in the volume of nuclease-free water to 10 μL used to elute the RNA from the column; such modifications were made to reduce the amount of PCR inhibitors passing through the column. RNA in the eluates were enumerated using Volcano 2nd Generation qPCR (V2G-qPCR)24, 31, 33, 34 for the N3 gene33 of SARS-CoV-2. Inhibition was evaluated by adding a known amount of HIV-1 viral RNA to the 10 μL of RNA. The Cq values from the samples were compared to the same HIV-1 spike into 10 μL of nuclease free water to measure the degree of inhibition. Very little to no inhibition was observed when comparing HIV-1 RNA target Cq values of samples to the water control.
2.4. Culturing C. auris from Wastewater
Alongside the filtration of wastewater samples, culturing methods had to be experimentally determined as there was limited research describing the culturing of C. auris from wastewater as of the summer of 2022. The positive control provided by the pathology laboratory was cultured on CHROMAgar to determine the phenotypic expression of C. auris on the media based upon color, which can be viewed within the supplemental text (Figure S-1). Previous work associated with the culturing of fecal coliform bacteria24, 31 was the basis of the approach for experimentally determining the conditions for growing C. auris. Initially, three conditions were experimentally tested by filtering known amounts of raw wastewater through disposable Supor membrane cups (0.45 μm pore size, Pall Corp.) according to standard methods for fecal coliform analysis (APHA D9222D, 200–). Dilutions used were 20 mL, 10 mL and 1 mL, supplemented with sterile PBS to a total volume of 25–30 mL to facilitate the distribution of particles across the membrane. Once all liquid had been passed through the Supor membranes, the filters were removed and immediately placed upon CHROMAgar media plates.17 Subsequent samples processed used only the 1 mL dilution of raw wastewater given the overgrowth of Candida spp. colonies on the CHROMAgar media.
All plates were incubated at 42 °C for 48-hours. At the end of the incubation period, plates were wrapped with Parafilm and transported to the CFAR laboratory on ice for further analysis. Upon their arrival at CFAR, individual colonies, depending on phenotypic color were re-streaked onto new CHROMAgar plates and sub-cultured for determining specificity of growth for Candida spp. Sub-cultured colonies were incubated at 37°C for another 48-hours. Images of the initial, and re-streaked colonies can be viewed in the Supplemental information (Figures S-5 to S-7). The re-cultured Candida spp. were then analyzed by qPCR as described above to confirm which color shades corresponded to C. auris.
2.5. Data Analysis and Acquisition
Clinical case data for C. auris and COVID-19 infected patients at the University’s hospital was acquired from the electronic health record for the date range January 1st, 2020 – January 8th, 2023 (UM IRB# 20210164). Admitted patients positive for C. auris were available on a daily basis (e.g., patient census). Wastewater qPCR results for C. auris were measured and quantified on a weekly frequency of collection from May 18, 2022 – September 21, 2022, wherein SARS-CoV-2 qPCR results correspond to weekly collections from September 30, 2020 – November 18, 2022. Weekly collections for time periods that overlap were splits from the same samples (one for C. auris and another for SARS-CoV-2 processing). Correlation assessments to determine the relationship between clinical cases and qPCR data, both Pearson (R) and Spearman (r), were performed with this data set and corresponded to the time frame for which data were available. Moreover, paired T-tests were utilized to assess the difference between the qPCR mean values of the wastewater collection sites. The statistical software package SPSS v28.0.0.0 was utilized for all analyses. Figures were created using Microsoft Excel.
3.0. Results and Discussion
Of the 19 WM06 samples analyzed for C. auris by qPCR from the hospital, 9 were positive, whereas only 3 (of 19) were positive from the CDWWTP site WC0Dc. Only one sample (of 19) was positive for both locations. The average levels of the C. auris positive samples collected from the hospital measured at 1.3 × 105 gc/L, which was one order of magnitude higher than that measured at the CDWWTP (1.9 × 104 gc/L). These differences, however, were not statistically significant via paired T-test (p=0.12) and can be viewed within Table S-2 within the supplemental information. These results suggest either dilution or degradation of the target by the time it reached the downstream collection location.
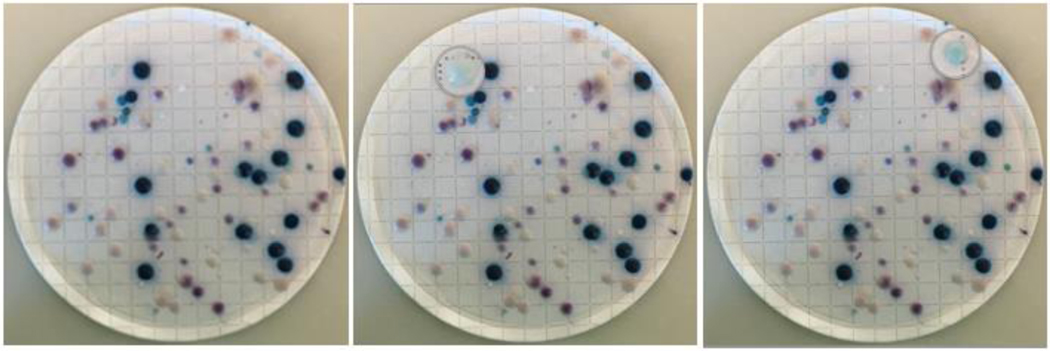
Culturing C. auris from wastewater to assess viability showed overgrowth of Candida colonies from the 20 mL and 10 mL conditions described above (Methods 2.3), the 1 mL condition was utilized and replicated. Within the supplemental text (Figure S-2 – S-7), images of colony growth on CHROMAgar media provide that multiple species of Candida grow in wastewater, and a very small fraction of those growths were ultimately C. auris (Figure 3). To determine the effective phenotypic classification of the CHROMAgar media, individual colonies of differing colors (i.e., white, navy, pink, purple) were re-streaked and incubated for another 48 hours to sub-culture larger quantities of CFU’s (colony forming units). The sub-cultures that underwent a second incubation time underwent the same DNA extraction method utilized for the qPCR analysis of the wastewater samples, including bead-bashing, and were assessed by the qPCR assay targeting C. auris. The percentage of Candida auris positive colonies was low relative to the negatives. Initially, identification of positives was based on the CHROMAgar manufacturer’s guide to evaluate teal-colored colonies. Since these were qPCR negative, two of each colored colony per primary plate were re-streaked to provide nucleic acids for confirmatory qPCR tests. Of these, colonies that appeared as a teal bullseye with a white halo were confirmed to be C. auris while all other colonies were qPCR negative. A representative example is provided in Figure 3 in which two of 90 (2.2%) total colonies were positive by qPCR for C. auris.
Figure 3:
Colonies of Candida spp. on CHROMAgar from 1 mL of wastewater collected on September 7, 2022, re-filtered after two weeks of hold time at 4 °C, and incubated at 37 °C for 48-hours. The left panel corresponds to the unaltered image. The middle and right panels show magnifications of light blue colonies that were positive for C. auris by qPCR.
A larger amplicon from the same target region was generated from two samples to further confirm by sequencing that the teal, haloed colonies were C. auris. Sequence alignment using ClustalW resulted in a 100% match between the samples and a confirmed C. auris sequence file deposited in GenBank at the NCBI (Table 1). Further experimentation to better define the relative abundance of C. auris to other Candida species within wastewater is needed to develop a more comprehensive understanding of the significance of detecting C. auris in wastewater. Expansion of sampling sites and temporal monitoring of C. auris levels may provide an improved knowledge base from which additional conclusions can be drawn. Additional methodology still needs to be assessed to define the Candida population found in wastewater and whether species other than C. auris, which may be more abundant, are of medical concern. Furthermore, the length of time C. auris remains viable in wastewater was not addressed in this study, so further research should be conducted to establish its persistence.
Table 1:
PCR primers and reporter probe used to specifically detect Candida auris. Reagents target the genomic region spanning the internal transcribed spacer 1, 28S rRNA, 5.8S rRNA and internal transcribed spacer 2.
| Assay | Primers/Probe Sequences | |
|---|---|---|
| qPCR Standards | Forward | GGATCTCTTGGTTCTCGCA |
| Reverse | GTAGGTGAACCTGCGGAAG | |
| qPCR | Forward | CGTGATGTCTTCTCACCAATCT |
| Reverse | TACCTGATTTGAGGCGACAAC | |
| Probe | 5HEX-TTTGTGAAT/ZEN/GCAACGCCACCGC-3IABkFQ | |
| Sequencing | Forward | TTTGCATACACACTGATTTGGA |
| Reverse | TACCTGATTTGAGGCGACAAC |
Overall, results show that we were able to expand workflows designed to evaluate SARS-CoV-2 to include C. auris. In this study we found that viable C. auris was detectable in wastewater from wastewater treatment plants using WBS approaches, which agrees with recently published literature.28, 29 Moreover, we have further documented that C. auris can be cultured from hospital wastewater, as suggested by Barber et al. and Rossi et al. from comparisons between wastewater collected at treatment plants with and without hospitals within their sewersheds.28, 29 Further research should be performed to assess whether differences in detected C. auris vary between geographical location and/or climate conditions due to the nature of C. auris’ thermo- and halotolerance as the current study’s data is captured from a different set of environmental conditions than those previously published.
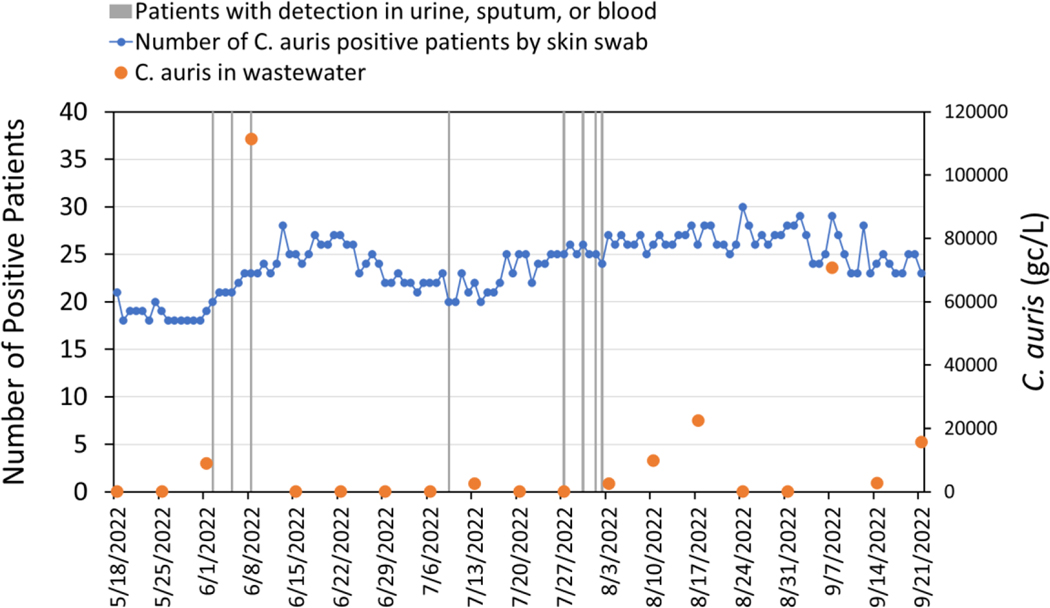
Our study further expands upon prior work by the inclusion of clinical C. auris patient data. The total number of colonized cases with C. auris in the hospital building during the targeted C. auris wastewater analysis period varied between 17 and 30 cases (Figure 1). Cases included patients positive by skin swab (using an axilla/groin composite swab as recommended by the CDC) plus an additional eight patients who also presented with C. auris in either urine, sputum, or blood samples (Table 2). qPCR detection of C. auris from the hospital wastewater during this time period varied, with qPCR results reporting as below detection limits (BDL) during most sampling weeks in spite of having patients admitted with known colonization. In two portions of the sampling period (June 1st, 2022, through June 8th, 2022, as well as July 27th, 2022, through August 17th, 2022), there was an increase in reported clinical cases of C. auris among those admitted to the hospital that somewhat followed the trend in wastewater detection. However, qPCR positives were only reported on 9 of the 19 weeks in which wastewater samples were assessed for C. auris.
Figure 1:
Time series plot of (left axis) daily clinical C. auris data for patients in the hospital as determined by skin swab (shown by blue circles) versus times when patients (n=1 for each vertical line) also tested positive in either urine, sputum or blood samples, and plot (right axis) for C. auris (gc/L) concentrations in wastewater samples.
Table 2:
Summary of average daily admitted patient census data based upon screening skin swab measurements and organism isolation from urine, sputum, and blood per date range, and sample type. Text in bold at the bottom of the table represent cumulative values for the entire period of record.
| Date Range | Average Daily Admitted Patient Census with Positive Skin Swab for C. auris of Specified Date Range | Number Detected In | ||
|---|---|---|---|---|
| Urine | Sputum | Blood | ||
| 5/18/20–2 – 5/31/2022 | 19 | 0 | 0 | 0 |
| 6/1/20–2 – 6/30/2022 | 24 | 1 | 0 | 2 |
| 7/1/20–2 – 7/31/2022 | 23 | 1 | 1 | 1 |
| 8/1/20–2 – 8/31/2022 | 27 | 0 | 0 | 2 |
| 9/1/20–2 – 9/21/2022 | 25 | 0 | 0 | 0 |
| 5/18/20–2 – 9/21/2022 | 24 | 2 | 1 | 5 |
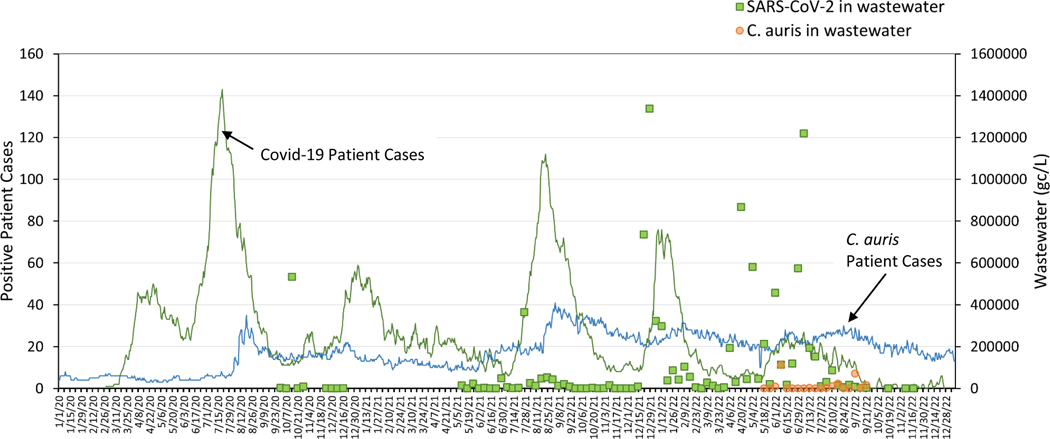
The SARS-CoV-2 RNA target in wastewater was also analyzed by qPCR for these same samples, and daily cases of COVID-19 were compiled from the hospital. A time series plot (Figure 2) illustrates the abundance of C. auris DNA and SARS-CoV-2 RNA from collected wastewater measured in genomic copies per liter (gc/L) as well as the trend in cases for both pathogens in the hospital. Although it is recognized that C. auris and SARS-CoV-2 are markedly different microbes, we believe that within a hospital setting there may be factors that may interlink transmission. For example, both C. auris and SARS-CoV-2 emerged simultaneously in our University hospital setting.35 There have been a number of outbreaks of C. auris reported in COVID-19 wards worldwide (and fungal pathogens overall).11, 12, 35, 36 While the exact biological mechanisms warrant further investigation (e.g., antibiotic over-use in COVID-19 disease driving microbiome changes, PPE misuse driving C. auris transmission events in COVID-19 wards, or in vivo biological interactions between the two organisms driving C. auris microbiome expansion), we believed it reasonable to correlate the two pathogens in wastewater given the expansion of both of these organisms during the pandemic time period.
Figure 2:
Time series plot of daily clinical data for C. auris and COVID-19 positive cases recorded in the University Hospital and quantified wastewater signal of C. auris and SARS-CoV-2 (gc/L), collected weekly. Gaps in wastewater data provide that no hospital sample was collected on that given date.
While it has been established that SARS-CoV-2 reported in a clinical setting mirrors the detectable concentration in wastewater, with variations of lag time22, 37, we found minimal and weak correlations between C. auris positive patients, by daily census, and corresponding wastewater concentrations. As wastewater was collected from a hospital collection site as well as the downstream WWTP, Pearson and Spearman correlations were performed between each sampling site and the daily census data to determine if a correlation existed similar to that of SARS-CoV-2 with WBS data22–24. Between sampling sites which compared the WWTP (WC0Dc) and hospital (WM06) there were insignificant correlations by both tests, [R = −0.068 (p = 0.781), and r = −0.054 (p = 0.827)], wherein the C. auris qPCR signal from either location had little to no relationship. Similarly, when comparing wastewater (either WC0Dc or WM06) against the daily census clinical data, results also showed insignificant correlations by Pearson and Spearman (p>0.69) (Table S-2).
These results illustrate that although C. auris may be detectable via wastewater, further research must be done regarding the utility of WBS approaches for correlating the abundance as a tool for clinical surveillance. The results generated by this study give evidence that Candida’s clinical abundance must be rather high to be detected in large quantities from wastewater, for accurate correlations to be determined. Future work should focus on increasing the sensitivity of C. auris detection in wastewater by considering large volume concentration methods38 and methods to amplify the C. auris signal upon culture by minimizing the growth of other Candida species. Reduction of interfering Candida species will permit for the filtration of larger volumes while isolating pure C. auris cultures.
4.0. Conclusions
The documentation of C. auris among communities is important because it can colonize patients and cause invasive infections, expresses resistance to antifungal agents when infection occurs, and it is difficult to detect and identify. The paucity of equipment to detect C. auris throughout the world is the main reason other methods of detection should be explored, such as wastewater. In this study workflows developed for SARS-CoV-2 virus quantification were extended to evaluate C. auris fungi levels in wastewater. Results show that C. auris could be detected using molecular methods and that C. auris remains viable in wastewater as it could be recovered using culture-based methods.
This is the first study to our knowledge that evaluates wastewater levels of C. auris from a single acute care facility with documented numbers of patients colonized or infected with C. auris. Additionally, this is the first time that wastewater levels at an acute care facility were compared to levels measured at the downstream WWTP. Though there were two periods during the sampling time frame in which there was a general trend between wastewater positivity and clinical infections with C. auris, there were no clear correlations with the total number of patients colonized. Similar results were observed for the SARS-CoV-2 hospital wastewater samples which also showed weak to negligible correlations with COVID-19 patients. The lack of correlation between C. auris patient census and wastewater levels can be due to variations in hospital practice with use of trash versus wastewater disposal and differing bioburdens of colonization in gut/skin of patients compared to SARS-CoV-2, although, there are data to hint at biologic mechanisms of gut microbiome expansion of fungal pathogens, such as C. auris, during a SARS-CoV-2 infection when compared with controls.39 The sporadic nature of the C. auris results in wastewater may also be associated with the fact that it is predominantly a skin pathogen. Within the UHealth hospital, there is no in-hospital laundry facility, so one of the plausible ways for this skin pathogen to enter the sewer system (via clothes washing) is not represented in this study. Thus, its inclusion in wastewater would be more sporadic and associated with inclusion of hand washing and bathing water from positive patients. Regardless of this more complex route of entering the wastewater, C. auris was still observed showing promise for its detection by WBS approaches. More research is needed to evaluate the mechanisms of C. auris entry into wastewater. Once in wastewater, the persistence of the DNA and culture-based signals should be explored further.
As we move forward from the COVID-19 pandemic, studies such as this aid in the narrative of using WBS for evaluating alternative pathogens of importance. Given C. auris’ impact – including cost – on hospital patient length of stay, further analysis should further evaluate its infectivity or transmission within a community.
Supplementary Material
Highlights.
C. auris isolated from wastewater (WW) using culture and PCR methods
First time comparisons made between hospital inpatient cases and wastewater
C. auris levels in WW were sporadic relative to number of hospital inpatients
C. auris levels were higher in hospital WW relative to community WW
C. auris shows promise for its detection through WW
Acknowledgements:
The research reported in this publication was supported by the National Institute on Drug Abuse of the National Institutes of Health (NIH) under Award Number U01DA053941 and the University of Miami Center for AIDS Research grant under award number P30AI073961. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Credit Author Statement
Kristina Babler: Methodology, Visualization, Formal Analysis, Writing – Original Draft, Writing-Review. Mark Sharkey: Conceptualization, Methodology, Writing – Review. Sebastian Arenas: Methodology. Ayaaz Amirali: Methodology. Cynthia Beaver: Methodology. Samuel Comerford: Methodology. Kenneth Goodman: Methodology. George Grills: Conceptualization, Methodology. Michelle Holung: Methodology, Conceptualization. Erin Kobetz: Methodology. Jennifer Laine: Methodology. Walter Lamar: Methodology. Christopher Mason: Conceptualization, Methodology, Supervision, Funding Acquisition. Darryl Pronty: Methodology. Brian D. Reding: Methodology. Stephan Schürer: Data Methodology, Funding Acquisition. Natasha Schaefer Solle: Methodology. Mario Stevenson: Resources. Dusica Vidović: Data Methodology. Helena Solo-Gabriele: Conceptualization, Methodology, Writing – Review, Supervision, Project Administration, Funding Acquisition. Bhavarth Shukla: Conceptualization, Methodology, Writing – Original Draft, Supervision.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5.0 References
- 1.Robert VA; Casadevall A, Vertebrate endothermy restricts most fungi as potential pathogens. J Infect Dis 2009, 200, (10), 1623–6. [DOI] [PubMed] [Google Scholar]
- 2.Jackson BR; Chow N; Forsberg K; Litvintseva AP; Lockhart SR; Welsh RM; Vallabhaneni S; Chiller T, On the Origins of a Species: What Might Explain the Rise of Candida auris? Journal of Fungi 2019, 5, (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A; Sharma C; Meis JF, Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 2017, 13, (5), e1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iguchi S; Itakura Y; Yoshida A; Kamada K; Mizushima R; Arai Y; Uzawa Y; Kikuchi K, Candida auris: A pathogen difficult to identify, treat, and eradicate and its characteristics in Japanese strains. J Infect Chemother 2019, 25, (10), 743–749. [DOI] [PubMed] [Google Scholar]
- 5.Kean R; Brown J; Gulmez D; Ware A; Ramage G, Candida auris: A Decade of Understanding of an Enigmatic Pathogenic Yeast. J Fungi (Basel) 2020, 6, (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira HC; Monteiro MC; Rossi SA; Peman J; Ruiz-Gaitan A; Mendes-Giannini MJS; Mellado E; Zaragoza O, Identification of Off-Patent Compounds That Present Antifungal Activity Against the Emerging Fungal Pathogen Candida auris. Front Cell Infect Microbiol 2019, 9, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Gaviria M; Martinez-Alvarez JA; Chavez-Santiago JO; Mora-Montes HM, Candida haemulonii Complex and Candida auris: Biology, Virulence Factors, Immune Response, and Multidrug Resistance. Infect Drug Resist 2023, 16, 1455–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora P; Singh P; Wang Y; Yadav A; Pawar K; Singh A; Padmavati G; Xu J; Chowdhary A, Environmental Isolation of Candida auris from the Coastal Wetlands of Andaman Islands, India. mBio 2021, 12, (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall A; Kontoyiannis DP; Robert V, Environmental Candida auris and the Global Warming Emergence Hypothesis. mBio 2021, 12, (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escandon P, Novel Environmental Niches for Candida auris: Isolation from a Coastal Habitat in Colombia. J Fungi (Basel) 2022, 8, (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casadevall A; Kontoyiannis DP; Robert V, On the Emergence of Candida auris: Climate Change, Azoles, Swamps, and Birds. mBio 2019, 10, (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti A; Sood P, On the emergence, spread and resistance of Candida auris: host, pathogen and environmental tipping points. J Med Microbiol 2021, 70, (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffery-Smith A; Taori SK; Schelenz S; Jeffery K; Johnson EM; Borman A; Candida auris Incident Management, T.; Manuel, R.; Brown, C. S., Candida auris: a Review of the Literature. Clin Microbiol Rev 2018, 31, (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welsh RM; Bentz ML; Shams A; Houston H; Lyons A; Rose LJ; Litvintseva AP, Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J Clin Microbiol 2017, 55, (10), 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(CDC), C. f. D. C. a. P., Antibiotic Resistance Threats in the United States. 2019.
- 16.Chowdhary A; Sharma C; Duggal S; Agarwal K; Prakash A; Singh PK; Jain S; Kathuria S; Randhawa HS; Hagen F; Meis JF, New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 2013, 19, (10), 1670–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MN; Shin JH; Sung H; Lee K; Kim EC; Ryoo N; Lee JS; Jung SI; Park KH; Kee SJ; Kim SH; Shin MG; Suh SP; Ryang DW, Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 2009, 48, (6), e57–61. [DOI] [PubMed] [Google Scholar]
- 18.Lee WG; Shin JH; Uh Y; Kang MG; Kim SH; Park KH; Jang HC, First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 2011, 49, (9), 3139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockhart SR; Etienne KA; Vallabhaneni S; Farooqi J; Chowdhary A; Govender NP; Colombo AL; Calvo B; Cuomo CA; Desjardins CA; Berkow EL; Castanheira M; Magobo RE; Jabeen K; Asghar RJ; Meis JF; Jackson B; Chiller T; Litvintseva AP, Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis 2017, 64, (2), 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh BJ; Shin JH; Kim MN; Sung H; Lee K; Joo MY; Shin MG; Suh SP; Ryang DW, Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol 2011, 49, (1), 98–102. [DOI] [PubMed] [Google Scholar]
- 21.Satoh K; Makimura K; Hasumi Y; Nishiyama Y; Uchida K; Yamaguchi H, Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009, 53, (1), 41–4. [DOI] [PubMed] [Google Scholar]
- 22.van Schalkwyk E; Mpembe RS; Thomas J; Shuping L; Ismail H; Lowman W; Karstaedt AS; Chibabhai V; Wadula J; Avenant T; Messina A; Govind CN; Moodley K; Dawood H; Ramjathan P; Govender NP; Germs SA, Epidemiologic Shift in Candidemia Driven by Candida auris, South Africa, 2016–2017(1). Emerg Infect Dis 2019, 25, (9), 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(CDC), C. f. D. C. a. P., Tracking Candida auris. 2022.
- 24.Kohlenberg A; Monnet DL; Plachouras D; Candida auris survey collaborative, g., Increasing number of cases and outbreaks caused by Candida auris in the EU/EEA, 2020 to 2021. Euro Surveill 2022, 27, (46). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prestel C; Anderson E; Forsberg K; Lyman M; de Perio MA; Kuhar D; Edwards K; Rivera M; Shugart A; Walters M; Dotson NQ, Candida auris Outbreak in a COVID-19 Specialty Care Unit - Florida, July-August 2020. MMWR Morb Mortal Wkly Rep 2021, 70, (2), 56–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briano F; Magnasco L; Sepulcri C; Dettori S; Dentone C; Mikulska M; Ball L; Vena A; Robba C; Patroniti N; Brunetti I; Gratarola A; D’Angelo R; Di Pilato V; Coppo E; Marchese A; Pelosi P; Giacobbe DR; Bassetti M, Candida auris Candidemia in Critically Ill, Colonized Patients: Cumulative Incidence and Risk Factors. Infect Dis Ther 2022, 11, (3), 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyman M; Forsberg K; Reuben J; Dang T; Free R; Seagle EE; Sexton DJ; Soda E; Jones H; Hawkins D; Anderson A; Bassett J; Lockhart SR; Merengwa E; Iyengar P; Jackson BR; Chiller T, Notes from the Field: Transmission of Pan-Resistant and Echinocandin-Resistant Candida auris in Health Care Facilities - Texas and the District of Columbia, January-April 2021. MMWR Morb Mortal Wkly Rep 2021, 70, (29), 1022–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forsberg K; Woodworth K; Walters M; Berkow EL; Jackson B; Chiller T; Vallabhaneni S, Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med Mycol 2019, 57, (1), 1–12. [DOI] [PubMed] [Google Scholar]
- 29.Meyer D; Martin EK; Madad S; Dhagat P; Nuzzo JB, Preparedness and response to an emerging health threat-Lessons learned from Candida auris outbreaks in the United States. Infect Control Hosp Epidemiol 2021, 42, (11), 1301–1306. [DOI] [PubMed] [Google Scholar]
- 30.Lockhart SR; Lyman MM; Sexton DJ, Tools for Detecting a “Superbug”: Updates on Candida auris Testing. J Clin Microbiol 2022, 60, (5), e0080821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kordalewska M; Perlin DS, Identification of Drug Resistant Candida auris. Front Microbiol 2019, 10, 1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plachouras D; Lotsch F; Kohlenberg A; Monnet DL; Candida auris survey collaborative, g., Candida auris: epidemiological situation, laboratory capacity and preparedness in the European Union and European Economic Area*, January 2018 to May 2019. Euro Surveill 2020, 25, (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyman M; Forsberg K; Sexton DJ; Chow NA; Lockhart SR; Jackson BR; Chiller T, Worsening Spread of Candida auris in the United States, 2019 to 2021. Ann Intern Med 2023, 176, (4), 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed W; Simpson SL; Bertsch PM; Bibby K; Bivins A; Blackall LL; Bofill-Mas S; Bosch A; Brandao J; Choi PM; Ciesielski M; Donner E; D’Souza N; Farnleitner AH; Gerrity D; Gonzalez R; Griffith JF; Gyawali P; Haas CN; Hamilton KA; Hapuarachchi HC; Harwood VJ; Haque R; Jackson G; Khan SJ; Khan W; Kitajima M; Korajkic A; La Rosa G; Layton BA; Lipp E; McLellan SL; McMinn B; Medema G; Metcalfe S; Meijer WG; Mueller JF; Murphy H; Naughton CC; Noble RT; Payyappat S; Petterson S; Pitkanen T; Rajal VB; Reyneke B; Roman FA Jr.; Rose JB; Rusinol M; Sadowsky MJ; Sala-Comorera L; Setoh YX; Sherchan SP; Sirikanchana K; Smith W; Steele JA; Sabburg R; Symonds EM; Thai P; Thomas KV; Tynan J; Toze S; Thompson J; Whiteley AS; Wong JCC; Sano D; Wuertz S; Xagoraraki I; Zhang Q; Zimmer-Faust AG; Shanks OC, Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci Total Environ 2022, 805, 149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X; Zhang S; Sherchan S; Orive G; Lertxundi U; Haramoto E; Honda R; Kumar M; Arora S; Kitajima M; Jiang G, Correlation between SARS-CoV-2 RNA concentration in wastewater and COVID-19 cases in community: A systematic review and meta-analysis. J Hazard Mater 2023, 441, 129848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F; Xiao A; Zhang J; Moniz K; Endo N; Armas F; Bonneau R; Brown MA; Bushman M; Chai PR; Duvallet C; Erickson TB; Foppe K; Ghaeli N; Gu X; Hanage WP; Huang KH; Lee WL; Matus M; McElroy KA; Nagler J; Rhode SF; Santillana M; Tucker JA; Wuertz S; Zhao S; Thompson J; Alm EJ, SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci Total Environ 2022, 805, 150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan Q; Babler KM; Sharkey ME; Amirali A; Beaver CC; Boone MM; Comerford S; Cooper D; Cortizas EM; Currall BB; Foox J; Grills GS; Kobetz E; Kumar N; Laine J; Lamar WE; Mantero AMA; Mason CE; Reding BD; Robertson M; Roca MA; Ryon K; Schurer SC; Shukla BS; Solle NS; Stevenson M; Tallon JJJ; Thomas C; Thomas T; Vidovic D; Williams SL; Yin X; Solo-Gabriele HM, Relationships between SARS-CoV-2 in Wastewater and COVID-19 Clinical Cases and Hospitalizations, with and without Normalization against Indicators of Human Waste. ACS ES&T 2022, (Water). [DOI] [PMC free article] [PubMed]
- 38.Tiwari A; Kurittu P; Al-Mustapha AI; Heljanko V; Johansson V; Thakali O; Mishra SK; Lehto KM; Lipponen A; Oikarinen S; Pitkanen T; WastPan Study G; Heikinheimo A, Wastewater surveillance of antibiotic-resistant bacterial pathogens: A systematic review. Front Microbiol 2022, 13, 977106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mataraci-Kara E; Ataman M; Yilmaz G; Ozbek-Celik B, Evaluation of antifungal and disinfectant-resistant Candida species isolated from hospital wastewater. Arch Microbiol 2020, 202, (9), 2543–2550. [DOI] [PubMed] [Google Scholar]
- 40.Sabino R; Verissimo C; Brandao J; Alves C; Parada H; Rosado L; Paixao E; Videira Z; Tendeiro T; Sampaio P; Pais C, Epidemiology of candidemia in oncology patients: a 6-year survey in a Portuguese central hospital. Med Mycol 2010, 48, (2), 346–54. [DOI] [PubMed] [Google Scholar]
- 41.Barber C; Crank K; Papp K; Innes GK; Schmitz BW; Chavez J; Rossi A; Gerrity D, Community-Scale Wastewater Surveillance of Candida auris during an Ongoing Outbreak in Southern Nevada. Environ Sci Technol 2023. [DOI] [PMC free article] [PubMed]
- 42.Rossi A; Chavez J; Iverson T; Hergert J; Oakeson K; LaCross N; Njoku C; Gorzalski A; Gerrity D, Candida auris Discovery through Community Wastewater Surveillance during Healthcare Outbreak, Nevada, USA, 2022. Emerg Infect Dis 2023, 29, (2), 422–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharkey ME; Babler KM; Amirali A; Grills GS; Kumar N; Laine J; Lamar WE; Mason CE; Reding BD; Schurer SC; Shukla B; Stevenson M; Vidovic D; Solo-Gabriele HM, First detection of the Monkeypox virus using wastewater-based surveillance in Miami-Dade County. Research Square 2022.
- 44.Sharkey ME; Kumar N; Mantero AMA; Babler KM; Boone MM; Cardentey Y; Cortizas EM; Grills GS; Herrin J; Kemper JM; Kenney R; Kobetz E; Laine J; Lamar WE; Mader CC; Mason CE; Quintero AZ; Reding BD; Roca MA; Ryon K; Solle NS; Schurer SC; Shukla B; Stevenson M; Stone T; Tallon JJ Jr.; Venkatapuram SS; Vidovic D; Williams SL; Young B; Solo-Gabriele HM, Lessons learned from SARS-CoV-2 measurements in wastewater. Sci Total Environ 2021, 798, 149177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lima A; Widen R; Vestal G; Uy D; Silbert S, A TaqMan Probe-Based Real-Time PCR Assay for the Rapid Identification of the Emerging Multidrug-Resistant Pathogen Candida auris on the BD Max System. J Clin Microbiol 2019, 57, (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Babler KM; Amirali A; Sharkey ME; Williams SL; Boone MM; Cosculluela GA; Currall BB; Grills GS; Laine J; Mason CE; Reding BD; Schurer SC; Stevenson M; Vidovic D; Solo-Gabriele HM, Comparison of Electronegative Filtration to Magnetic Bead-Based Concentration and V2G-qPCR to RT-qPCR for Quantifying Viral SARS-CoV-2 RNA from Wastewater. ACS ES&T Water 2022. [DOI] [PMC free article] [PubMed]
- 47.Babler KM; Sharkey ME; Abelson S; Amirali A; Benitez A; Cosculluela GA; Grills GS; Kumar N; Laine J; Lamar W; Lamm ED; Lyu J; Mason CE; McCabe PM; Raghavender J; Reding BD; Roca MA; Schurer SC; Stevenson M; Szeto A; Tallon JJ Jr.; Vidovic D; Zarnegarnia Y; Solo-Gabriele HM, Degradation rates influence the ability of composite samples to represent 24-hourly means of SARS-CoV-2 and other microbiological target measures in wastewater. Sci Total Environ 2023, 867, 161423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanson BM; Dinh AQ; Tran TT; Arenas S; Pronty D; Gershengorn HB; Ferreira T; Arias CA; Shukla BS, Candida auris Invasive Infections during a COVID-19 Case Surge. Antimicrob Agents Chemother 2021, 65, (10), e0114621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allaw F; Kara Zahreddine N; Ibrahim A; Tannous J; Taleb H; Bizri AR; Dbaibo G; Kanj SS, First Candida auris Outbreak during a COVID-19 Pandemic in a Tertiary-Care Center in Lebanon. Pathogens 2021, 10, (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janniger EJ; Kapila R, Public health issues with Candida auris in COVID-19 patients. World Med Health Policy 2021, 13, (4), 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.PAHO; WHO, Epidemiological Alert: Candida auris outbreaks in health care services in the context of the COVID-19 pandemic. In Organization, P. A. H., Ed. PAHO/WHO: Washington, D.C., 2021. [Google Scholar]
- 52.Solo-Gabriele HM; Kumar S; Abelson S; Penso J; Contreras J; Babler KM; Sharkey ME; Mantero AMA; Lamar WE; Tallon JJ Jr.; Kobetz E; Solle NS; Shukla BS; Kenney RJ; Mason CE; Schurer SC; Vidovic D; Williams SL; Grills GS; Jayaweera DT; Mirsaeidi M; Kumar N, Predicting COVID-19 cases using SARS-CoV-2 RNA in air, surface swab and wastewater samples. Sci Total Environ 2023, 857, (Pt 1), 159188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdelzaher AM; Solo-Gabriele HM; Palmer CJ; Scott TM, Simultaneous concentration of Enterococci and coliphage from marine waters using a dual layer filtration system. J Environ Qual 2009, 38, (6), 2468–73. [DOI] [PubMed] [Google Scholar]
- 54.Bonilla JA; Bonilla TD; Abdelzaher AM; Scott TM; Lukasik J; Solo-Gabriele HM; Palmer CJ, Quantification of Protozoa and Viruses from Small Water Volumes. Int J Environ Res Public Health 2015, 12, (7), 7118–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Francy DS; Stelzer EA; Brady AM; Huitger C; Bushon RN; Ip HS; Ware MW; Villegas EN; Gallardo V; Lindquist HD, Comparison of filters for concentrating microbial indicators and pathogens in lake water samples. Appl Environ Microbiol 2013, 79, (4), 1342–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haramoto E; Katayama H; Asami M; Akiba M, Development of a novel method for simultaneous concentration of viruses and protozoa from a single water sample. J Virol Methods 2012, 182, (1–2), 62–9. [DOI] [PubMed] [Google Scholar]
- 57.Hill VR; Polaczyk AL; Hahn D; Narayanan J; Cromeans TL; Roberts JM; Amburgey JE, Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl Environ Microbiol 2005, 71, (11), 6878–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMinn BR; Rhodes ER; Huff EM; Wanjugi P; Ware MM; Nappier SP; Cyterski M; Shanks OC; Oshima K; Korajkic A, Comparison of somatic and F+ coliphage enumeration methods with large volume surface water samples. J Virol Methods 2018, 261, 63–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morales-Morales HA; Vidal G; Olszewski J; Rock CM; Dasgupta D; Oshima KH; Smith GB, Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl Environ Microbiol 2003, 69, (7), 4098–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sassoubre LM; Love DC; Silverman AI; Nelson KL; Boehm AB, Comparison of enterovirus and adenovirus concentration and enumeration methods in seawater from Southern California, USA and Baja Malibu, Mexico. J Water Health 2012, 10, (3), 419–30. [DOI] [PubMed] [Google Scholar]
- 61.Zuo T; Zhan H; Zhang F; Liu Q; Tso EYK; Lui GCY; Chen N; Li A; Lu W; Chan FKL; Chan PKS; Ng SC, Alterations in Fecal Fungal Microbiome of Patients With COVID-19 During Time of Hospitalization until Discharge. Gastroenterology 2020, 159, (4), 1302–1310 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.