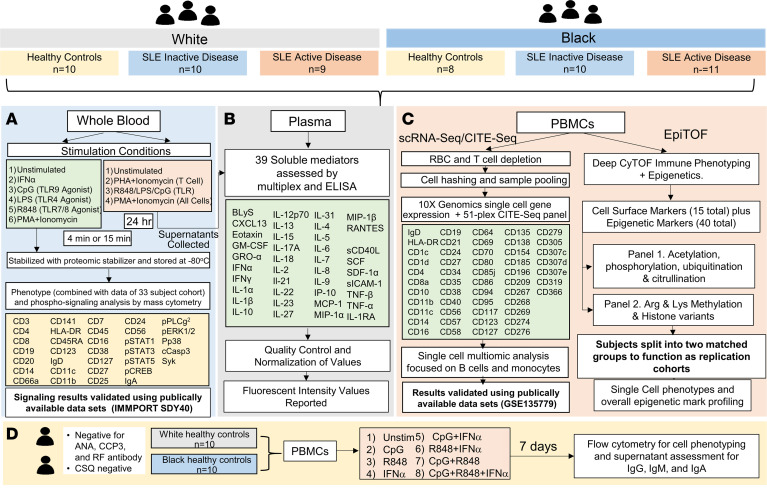
Figure 1. Overview of analysis workflow to catalog ancestry-associated differences in immune phenotypes.
A discovery cohort was used for a multiomic systems immunology analysis. Significant results were independently validated either by reusing publicly available data sets or by splitting individuals into 2 matched groups for replication (bold). First, 58 samples, including healthy controls, SLE INACT, and SLE ACT, who self-reported as White or Black, were matched by age, ancestry, and sex. (A) Whole blood was collected, left unstimulated or stimulated, and used for immunophenotyping by mass cytometry and signaling analysis by phospho-CyTOF. Similar data from an independently collected and analyzed set of samples, a larger combined analysis of 33 participants, were used to increase the power. (B) Plasma and supernatants collected after overnight stimulation of whole blood were used to assess 39 different soluble mediators using multiplex bead-based assays and ELISAs. (C) PBMCs were used for droplet-based scRNA-Seq and EpiTOF. For single-cell transcriptomics, PBMCs were washed, depleted of red blood cells and T cells using CD2 depletion to enrich for non–T cell populations, and stained with a 51-plex CITE-Seq panel for dual transcript and protein expression using the 10x Genomics 3′ single-cell droplet methods. These variables were utilized to delineate specific cell lineages, activation, and regulatory markers. (D) PBMCs from healthy controls with no autoimmune disease manifestations who self-reported as White or Black were stimulated for 7 days with IFN-α, TLR7/8, or TLR9 agonists, alone or in combination, to assess immune composition and antibody production by flow cytometry and ELISA, respectively. ANA, antinuclear antibody; CCP, cyclic citrullinated peptide; CSQ, connective tissue screening questionnaire; EpiTOF, CyTOF immune phenotyping with epigenetics; scRNA-Seq, sincle-cell RNA sequencing; MCP-1, monocyte chemoattractant protein-1; PHA, phytohemagglutinin; p-STAT, phosphorylated STAT; RF, rheumatoid factor; SCF, stem cell factor.