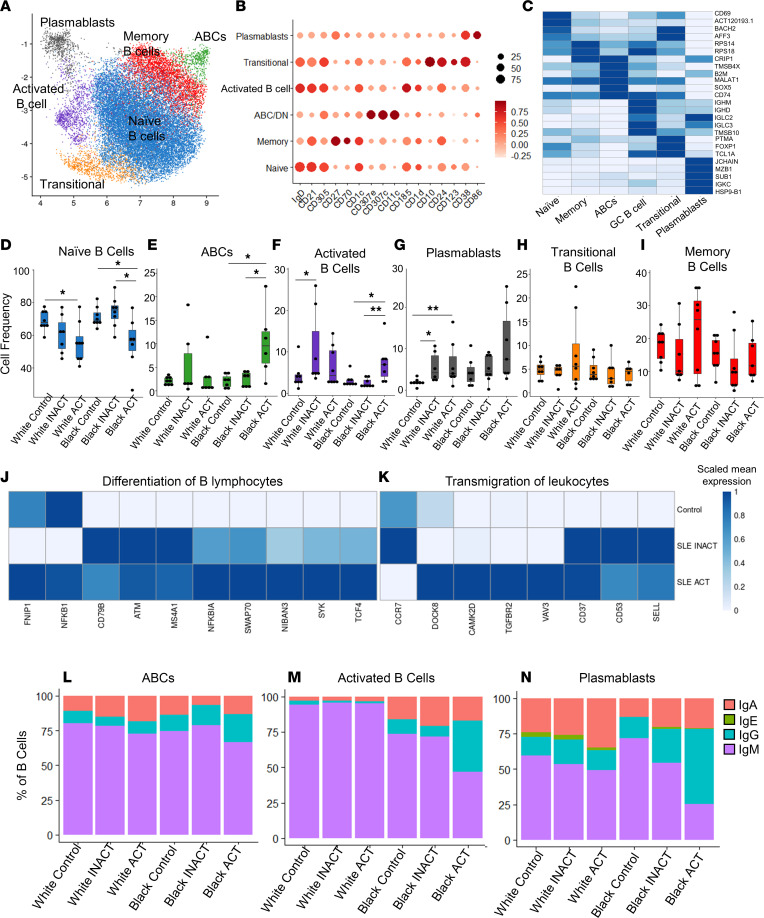
Figure 4. Alterations in the genomic landscape of patients with SLE ACT reveal greater IgG levels in Black patients.
PBMCs from 46 controls and patients with SLE were CD2-depleted, followed by droplet-based scRNA-Seq using 10x Genomics. (A) UMAP plot representing the 6 B cell clusters across all samples. The putative identity of each cluster was assigned using gene expression and protein expression from CITE-Seq. (B) Dot plot representing expression values of selected proteins assessed by CITE-Seq and (C) heatmap representing gene expression values of selected genes across each cluster used for cluster annotation. Dot size represents the percentage of cells expressing the marker of interest. Color intensity indicates the mean expression within expressing cells. (D–I) Box plots comparing the proportion (mean ± SD) of each cell type cluster across the disease groups and ancestries for (D) naive B cells, (E) age-associated B cells (ABCs), (F) activated B cells, (G) plasmablasts, (H) transitional B cells, and (I) memory B cells as defined by scRNA-Seq. Box plots show the interquartile range (box), median (line), and minimum and maximum (whiskers). Ingenuity Pathway Analysis (IPA; QIAGEN) of differentially expressed genes identified differences in (J) differentiation of B cells and (K) transmigration genes of naive B cells between patients with SLE and controls. Heatmaps shows scaled mean expression of genes in each pathway. (L–N) The percentages of B cells with high gene expression of class-switched IgA, IgE, IgG, or IgM are shown by B cell subset in (L) ABCs, (M) activated B cells, and (N) plasma cells. P values were calculated using pairwise Wilcoxon’s rank-sum tests between disease groups with Benjamini-Hochberg correction for multiple comparisons. *P < 0.05, and **P < 0.01.