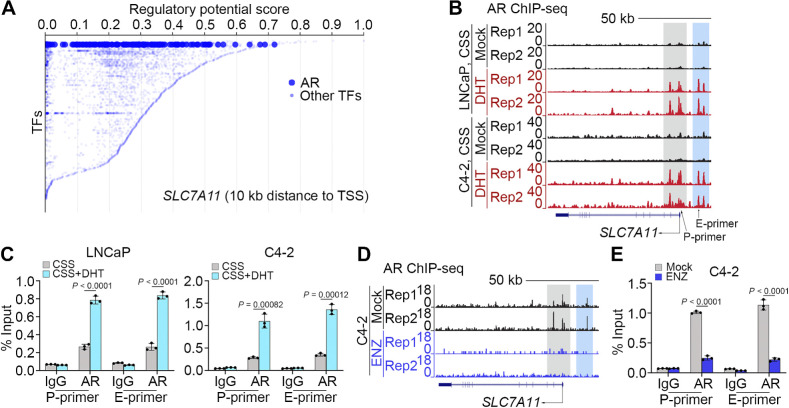
Figure 3.
AR promotes SLC7A11 expression at transcriptional level. A, Meta-analysis of ChIP data of transcription factors that potentially bind to the SLC7A11 gene locus using the online-based platform Cistrome software (http://dbtoolkit.cistrome.org). B and C, UCSC screenshot of AR ChIP-Seq showing AR occupancy at the SLC7A11 loci in LNCaP and C4–2 cells cultured in CSS medium and treated with or without DHT (B), and qChIP-PCR analysis of AR binding at the SLC7A11 promoter and enhancer regions in LNCaP and C4–2 (C) cells with the same treatment as in B. D and E, AR ChIP-Seq occupancy profiles at the SLC7A11 loci in C4–2 cells treated with or without ENZ (D). qChIP-PCR confirming AR binding at the SLC7A11 promoter and enhancer region was abolished in C4–2 cells (E) with the same treatment as in D.